Session Information
Date: Sunday, October 26, 2025
Title: (0430–0469) Rheumatoid Arthritis – Diagnosis, Manifestations, and Outcomes Poster I
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: Early diagnosis of rheumatoid arthritis (RA) enables prompt intervention, significantly mitigating risk for disease progression, joint damage, and associated morbidity. Conventional biomarkers, anti-CCP and rheumatoid factor (RF), have high specificity but limited sensitivity. Novel biomarkers such as RA33 (heterogeneous nuclear ribonucleoprotein A2/B1) and PAD4 (peptidyl arginine deiminase 4) antibodies have demonstrated diagnostic potential, though their combined utility remains unclear. This study evaluates the performance of novel biomarkers individually and in combination with traditional markers using machine learning algorithms.
Methods: A total of 760 subjects were enrolled, including 350 RA patients meeting 2010 ACR criteria (205 seropositive, 145 seronegative), 215 apparently healthy volunteers (AHV), and 195 disease controls (107 SLE meeting 1997 ACR criteria, 43 primary Sjögrens disease, and 45 psoriatic arthritis [PsA]). Serum samples were tested for anti-CCP IgG, RF IgM and IgA, RA33 (IgA, IgG, IgM), PAD4 (IgA, IgG), anti-SSB/La, and anti-Smith. Diagnostic cutoffs for anti-CCP and RF followed manufacturer recommendations; novel biomarker cutoffs were determined based on at least 95% specificity vs. AHVs. We evaluated multiple machine learning models and biomarker combinations using 10-fold cross-validation and independent test validation. Logistic regression models were built for individual conventional markers to compare against a Gradient Boosted Tree (GBT) classifier, which was ultimately selected based on superior AUC performance. Sensitivity and specificity were calculated at the optimal F1-maximized threshold.
Results: The RA cohort was predominantly female (77%), white (68%), with a mean age of 51 years (SD = 15) and a mean disease duration of 9 years (SD = 9) (Table 1). Anti-CCP was highly specific (99–100%) and RF IgM also showed high specificity (91–98%) (Table 2). RA33 markers were generally specific (71–98%), with RA33 IgG showing the highest sensitivity among them (13% in seronegative RA). PAD4 markers also had high specificity (92–98%), with PAD4 IgG demonstrating the highest sensitivity overall (44% in seropositive RA). PAD4 IgG had the highest positive likelihood ratio (+LR = 20.43), followed by PAD4 IgA (4.57), RA33 IgG (2.41), RA33 IgM (1.77), and RA33 IgA (1.55). The GBT classifier combining conventional and novel biomarkers (RA33s, PAD4s, anti-SSB La, Smith) achieved an AUC of 0.92 in both cross-validation and test sets (Figure 1A-B). The algorithm was 86% sensitive and 86% specific in cross-validation, and 81% sensitive and 90% specific in the test set. The GBT algorithm identified an additional 30% of RA cases in cross-validation (Figure 1C) and 29% in the test set (Figure 1D) that were missed by conventional biomarkers alone, leaving only 11–16% of RA patients undetected by any marker.
Conclusion: Integrating conventional biomarkers with novel RA33 and PAD4 antibodies through machine learning improves RA diagnostic accuracy, capturing ~30% of cases missed by conventional biomarkers alone. This enhanced diagnostic performance highlights the potential of advanced classifiers for earlier identification and timely management of RA.
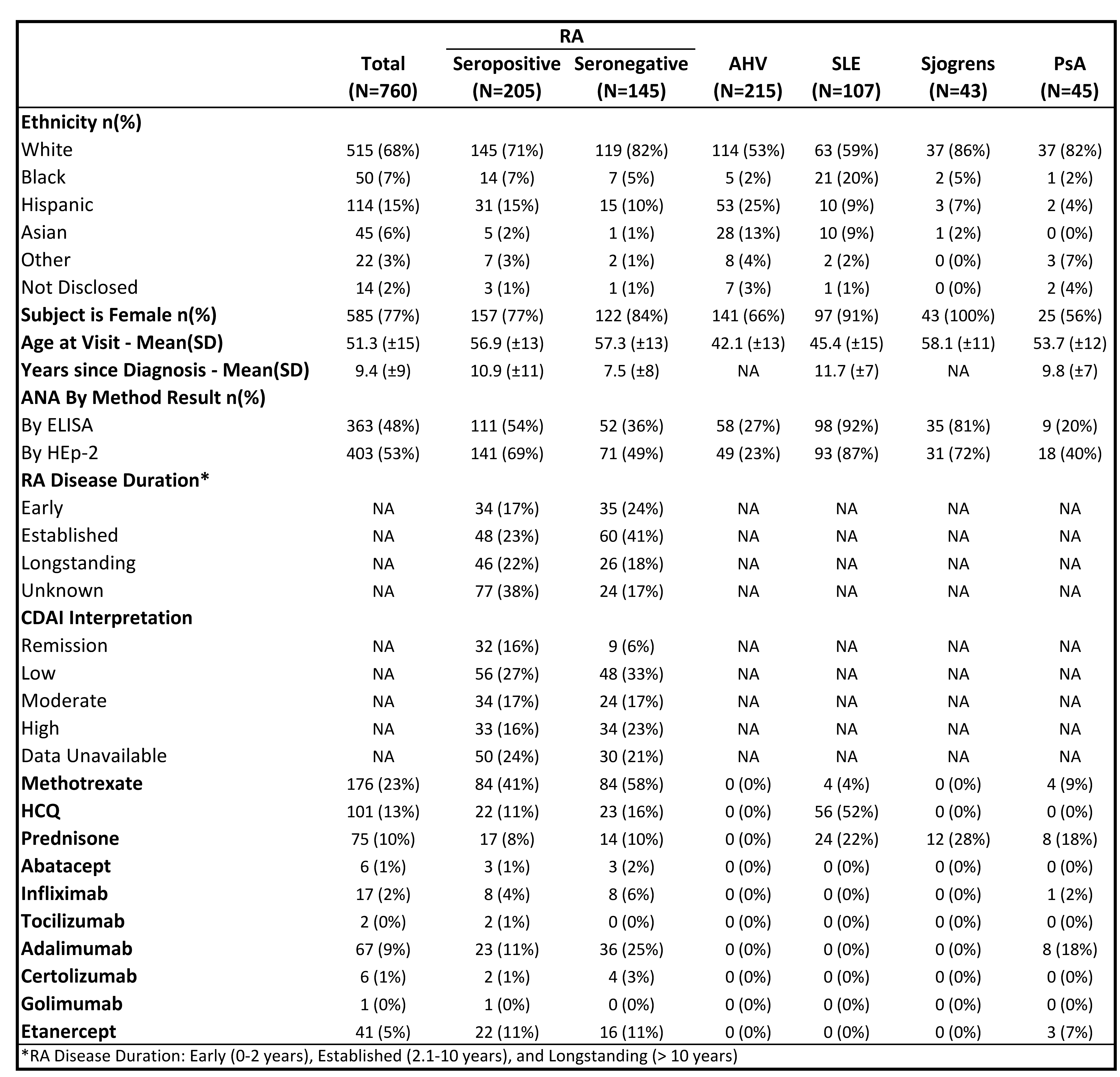 Table 1. Demographic characteristics
Table 1. Demographic characteristics
.jpg) Table 2. Performance characteristics
Table 2. Performance characteristics
.jpg) Figure 1. ROC analysis comparing algorithm performance to conventional biomarkers and sensitivity overlap analysis
Figure 1. ROC analysis comparing algorithm performance to conventional biomarkers and sensitivity overlap analysis
To cite this abstract in AMA style:
Kyttaris V, Taghavi S, Nagle C, Schleif C, Partain B, O'Malley T. A Machine Learning Classifier Leveraging Anti-RA33, Anti-PAD4, and Conventional Biomarkers Enhances Diagnostic Performance Characteristics for RA [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/a-machine-learning-classifier-leveraging-anti-ra33-anti-pad4-and-conventional-biomarkers-enhances-diagnostic-performance-characteristics-for-ra/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/a-machine-learning-classifier-leveraging-anti-ra33-anti-pad4-and-conventional-biomarkers-enhances-diagnostic-performance-characteristics-for-ra/
