Session Information
Date: Sunday, October 26, 2025
Title: (0098–0114) Spondyloarthritis Including Psoriatic Arthritis – Basic Science Poster
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: TNF inhibitors (TNFi) have proven efficacy in psoriatic arthritis (PsA) and are commonly prescribed as first line targeted therapy in PsA. However, response to TNFi is unpredictable and many patients fail to respond adequately, with a substantial minority not responding at all. Thus, there is an urgent need for biomarkers that can predict treatment response. Epigenetic chromosome conformation signatures (CCS) using EpiSwitch™ technology have shown predictive potential in various cancers and in predicting a methotrexate response in patients with rheumatoid arthritis [1] but have not been studied in PsA. This work aimed to identify predictive biomarkers of therapeutic response to adalimumab in patients with PsA using baseline peripheral blood CCS.
Methods: Baseline whole blood samples were used from participants in the OPAL Broaden phase III trial (NCT01877668) who received the TNFi adalimumab in the active comparator arm [2]. Response was defined using ACR response at 3 months. Baseline samples and response data were available for 91 participants, with 11 defined as extreme good responders (achieving at least an ACR70 response) and 11 as extreme non-responders (failing to reach at least ACR20 response) selected for the discovery set. Baseline peripheral blood samples underwent genome-wide chromosomal loop screening using the EpiSwitch™ Explorer microarray (Oxford Biodynamics), which consists of ~106 chromatin loops probes. Differential chromatin loops were identified and translated into qPCR assays for further feature reduction and signature refinement using penalised logistic regression.
Results: EpiSwitch™ Explorer microarray analysis of 22 (8 females, 14 males) extreme responders/non-responders, after correcting for biological sex, identified 2,291 chromatin loops (fold change >1.1, p-value < 0.05). Feature reduction narrowed these down to 50 loops, which were further analysed by qPCR in extreme responders/non-responders, as well as in moderate responders (achieving ACR50 but not ACR70 response), and partial responders (achieving ACR20 but not ACR50 response). 30 loops successfully passed the qPCR process. From these, a predictive model for treatment response containing 12 chromatin loop markers was developed, achieving an accuracy of 96.4% and an AUC of 0.93 (Figure 1).
Conclusion: A 12-loop chromosome conformation signature was modelled as a potential biomarker for predicting the therapeutic response to adalimumab in PsA patients in a phase III trial. This model demonstrated 96.4% accuracy and shows promise in correctly identifying not only extreme responders and non-responders, but also moderate/partial responders, highlighting its potential to help select the most effective personalised treatment for every individual. Further validation in an independent cohort is being planned to confirm the accuracy and reliability of these 12 loops as predictive biomarkers for TNFi response in PsA.References[1] Carini, C. et al. J Transl Med 2018;16. [2] Mease, P. et al. NEJM 2017; 377:1537–50.
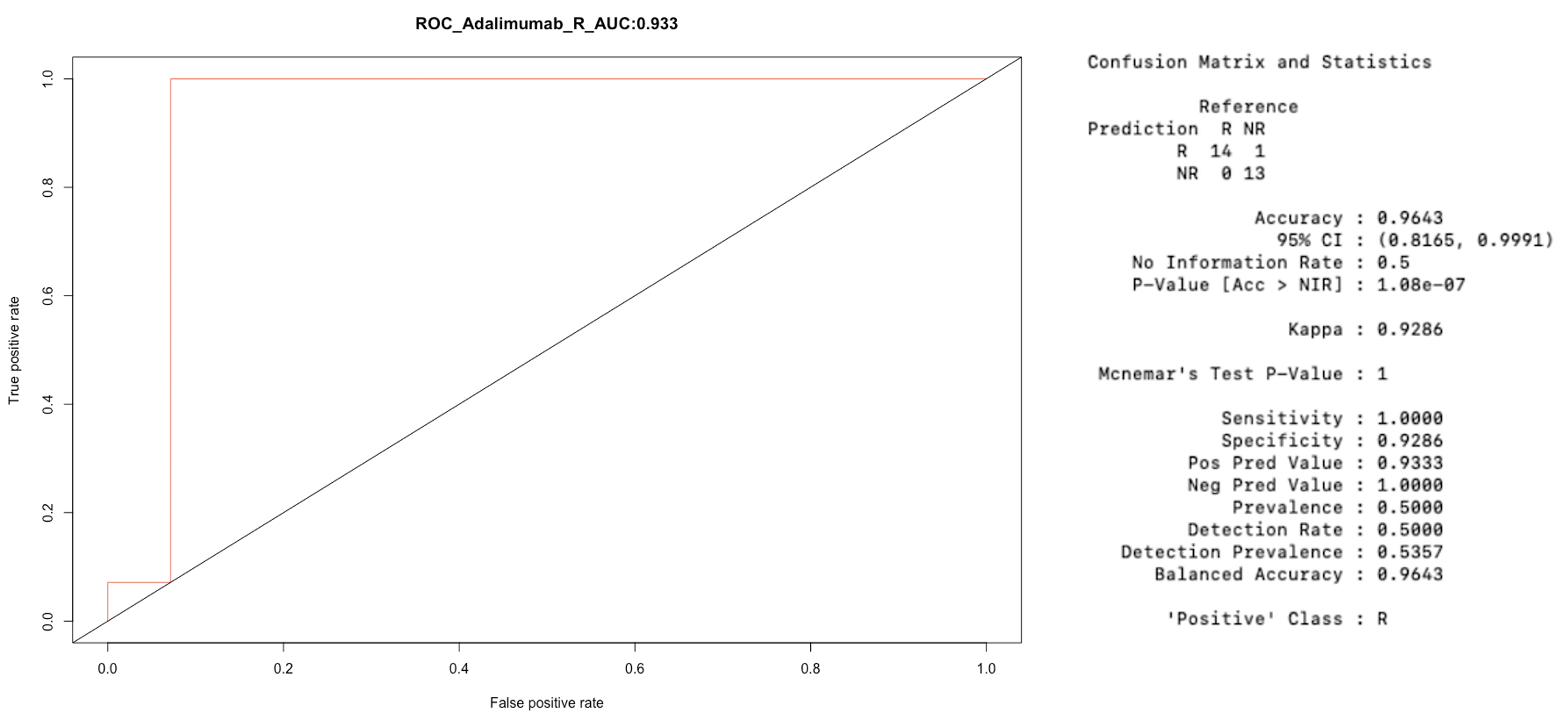 Figure 1: ROC curve displaying the performance of the selected 12-loop signature for PsA responders and non-responders to adalimumab, along with associated statistical matrix.
Figure 1: ROC curve displaying the performance of the selected 12-loop signature for PsA responders and non-responders to adalimumab, along with associated statistical matrix.
To cite this abstract in AMA style:
Riedlova P, Hunter E, Huppertz C, de Wit M, Pennington S, FitzGerald O, Schwenk J, Lories R, Goodyear C, Siebert S. Predicting Response To Adalimumab In Patients With Psoriatic Arthritis Using Epigenetic Chromosome Conformation Signatures [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/predicting-response-to-adalimumab-in-patients-with-psoriatic-arthritis-using-epigenetic-chromosome-conformation-signatures/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/predicting-response-to-adalimumab-in-patients-with-psoriatic-arthritis-using-epigenetic-chromosome-conformation-signatures/
