Session Information
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: IgA vasculitis (IgAV) is a systemic small-vessel vasculitis characterized by immune complex deposition containing aberrantly glycosylated IgA1. While its clinical manifestations are well defined [1], the molecular mechanisms driving disease onset and progression remain poorly understood. Diagnosis still relies on invasive tissue biopsies, underscoring the need for molecular biomarkers to support non-invasive alternatives. Although B cells are central to IgA biology, their role in IgAV pathogenesis remains unclear [2], particularly regarding the regulation of IgA glycosylation [3]. Here, we present the first single-cell RNA sequencing (scRNA-seq) analysis of peripheral B cells in IgAV, with the aim to identify cell subpopulations presenting transcriptional alterations associated with the disease’s ethiopathogenesis.
Methods: For this study we recruited 12 Caucasian IgAV patients in the acute phase of the disease and 12 healthy individuals matched by sex, age, and ethnicity to the patients. From the extracted peripheral blood, mononuclear cells were isolated using a density gradient with Ficol and purified the CD19+ B Cells using a magnetic separation kit using MACS® Technology. Single-cell separation and library preparation was carried out using the Chromium platform (10X technologies) and Illumina library preparation kits. Transcriptomic sequencing was carried out on a Novaseq X plus instrument. Raw sequencing data was preprocessed with cellranger and its outputs were analysed with Scanpy and Seurat.
Results: Interestingly, our results reveal for the first time a B cell subpopulation significantly enriched in IgAV patients compared to healthy controls (Fig. 1A). This subset, corresponding to Leiden cluster 6, showed a marked overrepresentation in the IgAV group, as illustrated by both the cluster-wise proportion comparison (Fig. 1B) and the heatmap of cluster proportions (Fig. 1C). Of note, differential gene expression analysis of this population revealed dysregulation of key genes involved in O-glycosylation pathways (Fig. 2), including known regulators of IgA glycosylation. These findings suggest that this B cell subset may contribute to IgAV pathogenesis through the production of aberrantly glycosylated IgA, highlighting its potential relevance as both a mechanistic driver and a biomarker of disease.
Conclusion: Our results reveal, for the first time, a B cell subpopulation with dysregulated glycosylation genes characteristic of IgAV, highlighting its potential as a disease-specific biomarker and a contributor to pathogenic IgA production.References:[1] Arthritis Rheum. 2013 Jan;65(1):1-11. [2] Front Immunol. 2022 Oct 3:13:921864. [3] Semin Nephrol. 2024 Sep;44(5):151571.Funding: European Union FEDER funds and “Fondo de Investigaciones Sanitarias” from “Instituto de Salud Carlos III” (ISCIII, Health Ministry, Spain), (PI21/00042 and PI24/00382 ). JCBL: PFIS program fellowship from the ISCIII, co-funded by the European Social Fund (`Investing in your future´), (FI22/00020), M-AES mobility grant (MV24/00004). RL-M: Miguel Servet type II program fellowship from the ISCIII, co-funded by ESF (“Investing in your future” (CPII21/00004).
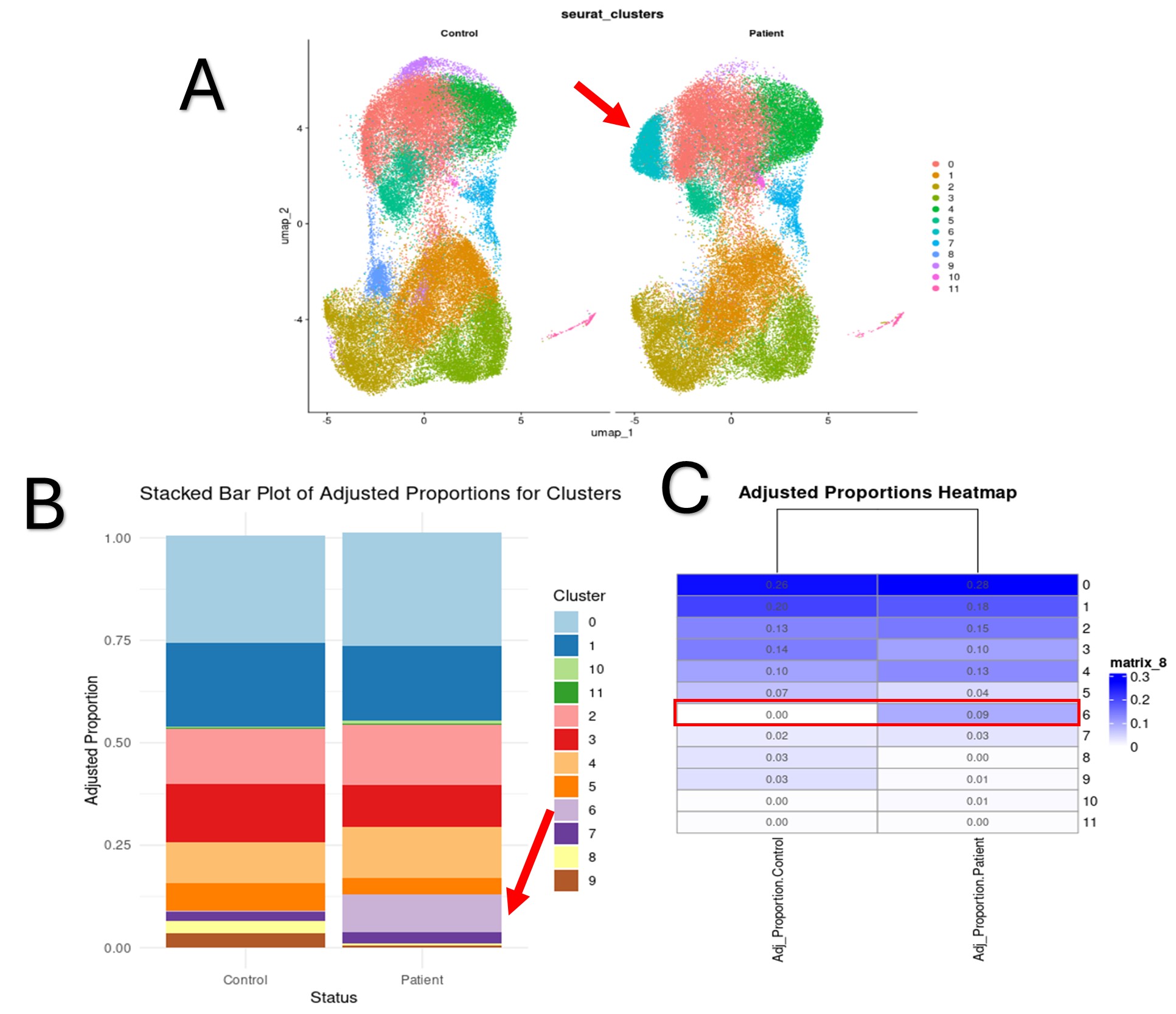 Figure 1. A) UMAP visualization of B cell subpopulations identified by Leiden clustering in IgAV patients and healthy controls. The red arrow indicates the cluster of interest (Leiden cluster 6), which is enriched in IgAV samples. B) Stacked bar plot showing the relative proportions of B cell clusters in each group. Leiden cluster 6, highlighted with a red arrow and box, is overrepresented in IgAV patients. C) Heatmap summarizing B cell cluster proportions across individual samples, further illustrating the expansion of Leiden cluster 6 in IgAV.
Figure 1. A) UMAP visualization of B cell subpopulations identified by Leiden clustering in IgAV patients and healthy controls. The red arrow indicates the cluster of interest (Leiden cluster 6), which is enriched in IgAV samples. B) Stacked bar plot showing the relative proportions of B cell clusters in each group. Leiden cluster 6, highlighted with a red arrow and box, is overrepresented in IgAV patients. C) Heatmap summarizing B cell cluster proportions across individual samples, further illustrating the expansion of Leiden cluster 6 in IgAV.
.jpg) Figure 2. Volcano plot of differential gene expression from the B cell subpopulation identified in Leiden cluster 6. Genes highlighted in red are associated with glycosylation pathways implicated in the production of aberrantly glycosylated IgA. Dotted line represents a 0.05 p-value threshold.
Figure 2. Volcano plot of differential gene expression from the B cell subpopulation identified in Leiden cluster 6. Genes highlighted in red are associated with glycosylation pathways implicated in the production of aberrantly glycosylated IgA. Dotted line represents a 0.05 p-value threshold.
To cite this abstract in AMA style:
Batista-Liz J, Sebastián Mora-Gil M, Troendle E, Cappa O, Leonardo M, Peñalba A, mARTIN pENAGOS L, Gabrie-Rodriguez L, Gálvez Sánchez R, Callejas J, Sevilla B, de Argila D, Vicente-Rabaneda E, Castañeda S, Simpson D, Blanco R, Pulito Cueto V, Lopez Mejias R. A Novel B Cell Subpopulation Characteristic of IgA Vasculitis Identified by Single-Cell RNA Sequencing [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/a-novel-b-cell-subpopulation-characteristic-of-iga-vasculitis-identified-by-single-cell-rna-sequencing/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/a-novel-b-cell-subpopulation-characteristic-of-iga-vasculitis-identified-by-single-cell-rna-sequencing/
