Session Information
Session Type: Abstract Session
Session Time: 1:00PM-2:30PM
Background/Purpose: The complement system is a major component of innate immunity and plays a vital role in experimental models of autoimmune disease pathogenesis. In patients with rheumatoid arthritis (RA), complement activation proteins generated in the synovium can interact with macrophages, promoting inflammation and disrupting tissue homeostasis for RA. However, it remains to be determined how specific complement components directly modulate cell-type functions. Understanding this may generate alternative therapeutic targets for RA.
Methods: We have generated a comprehensive RA synovial single-cell atlas and classified the spectrum of RA biopsies into six tissue phenotypes (Zhang, et al, Nature, 2023)1. In this current study, we linked single-cell transcriptomic expression of complement activation pathway components and receptors for effector proteins with cell-type heterogeneity and tested significant associations with different tissue types. Next, we characterized myeloid cell differentiation and aligned specific complement gene expression to myeloid functional states. Further, we established an in vitro co-culture system to evaluate the impacts of complement inhibitors on the transcriptional phenotype of macrophages.
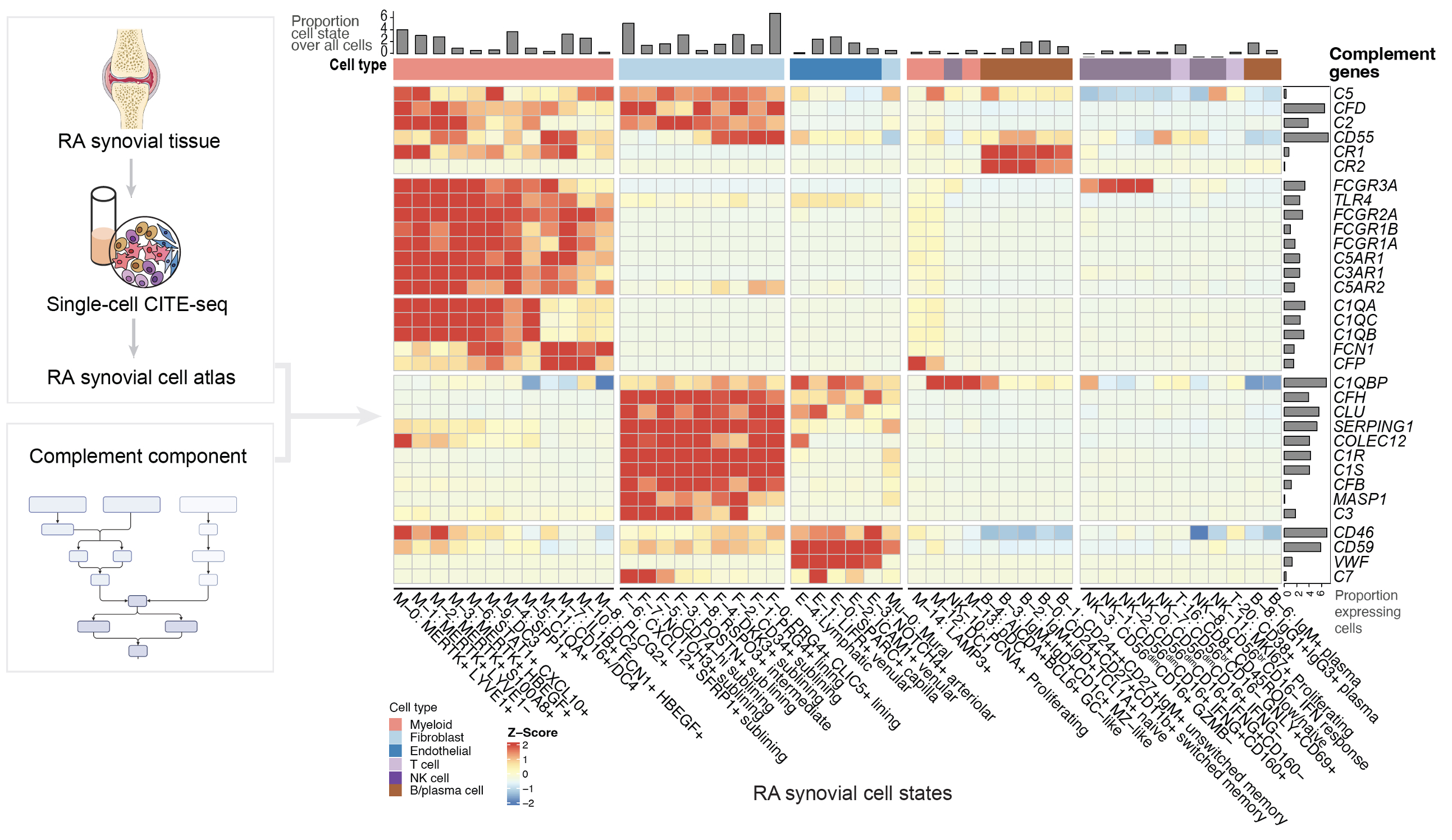
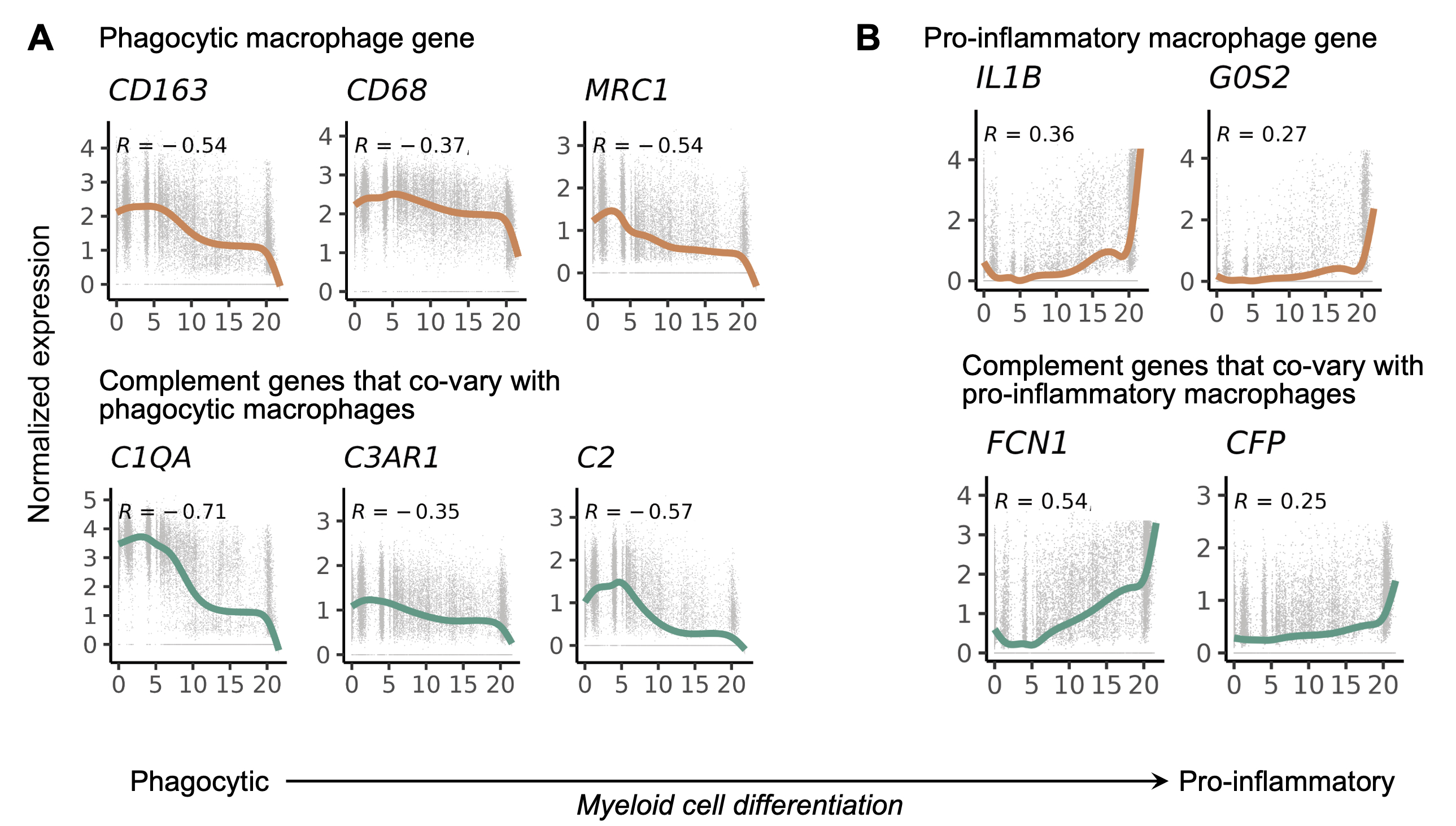
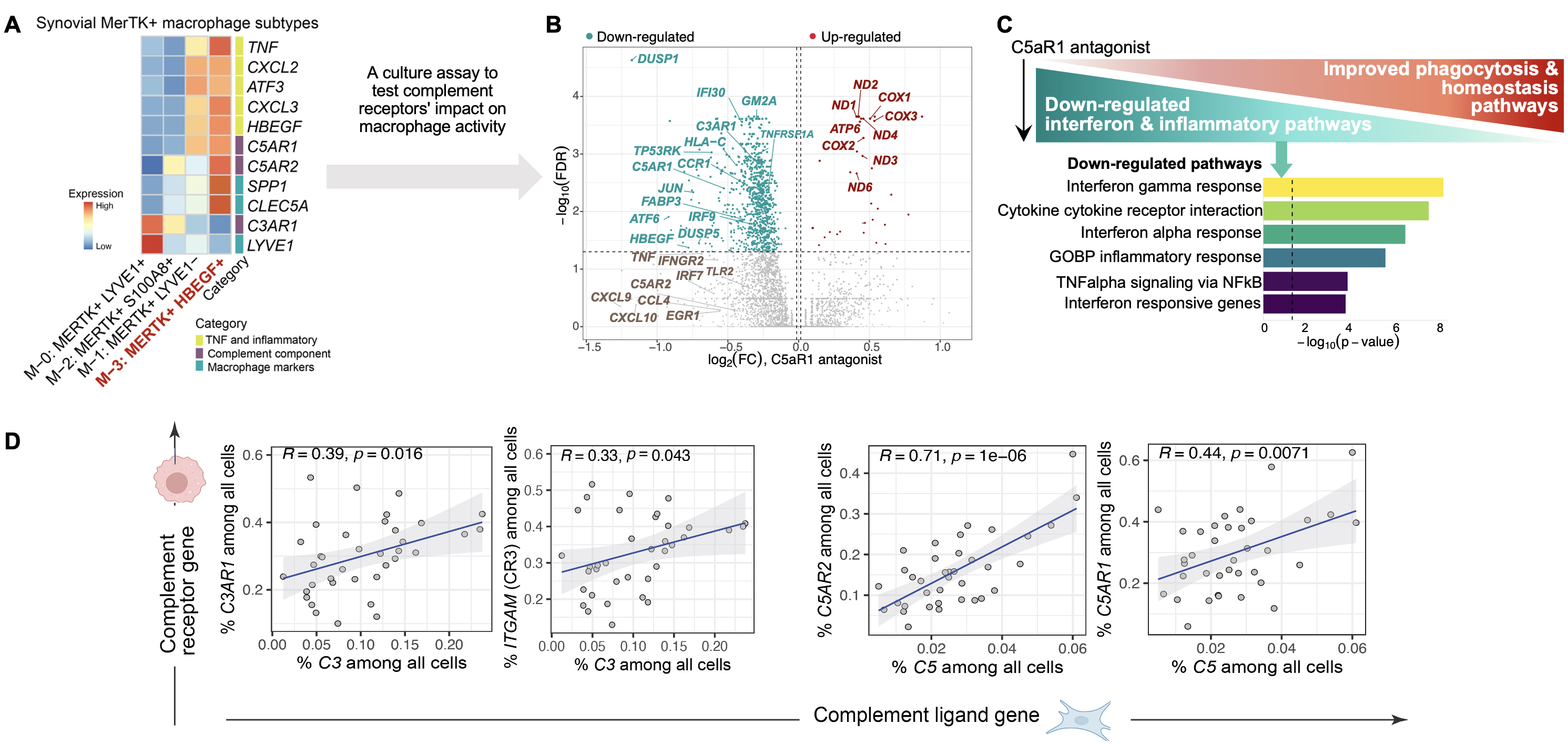
Results: We generated a complement cellular graph characterizing interactions between complement components and cell-type patterns in RA synovium. The complement components present unexpected distinct transcriptomic expression across cell types (Fig. 1). Within myeloid cells, we identified a myeloid cell differentiation axis revealing that complement components C1QA-B, C3AR1, and C2 expressions correlated with anti-inflammatory/phagocytic functions; while complement components FCN1 and CFP expression correlated with pro-inflammatory function (Fig. 2). Further, we identified a particular subtype, MERTK+HBEGF+ tissue macrophage, which is enriched in lymphocyte-low tissues and marked by the pro-inflammatory complement 5 receptor C5AR1, and inflammatory TNF signaling pathway-associated signatures like TNF, CXCL2, and CXCL3 (Fig. 3A). Intriguingly, we found that the addition of an inhibitor of C5aR1 suppressed inflammatory and interferon responses, while upregulating tissue protective phagocytic programs (Fig. 3B-C). Moreover, we revealed that the abundance of complement-dependent receptor-ligand (C5AR-C5 and C3AR1-C3) are correlated across RA patients (Fig. 3D), suggesting potential complement pathway crosstalk that impacts synovial macrophages and fibroblasts.
Conclusion: Through systematically aligning complement pathways with synovial heterogeneity, we found that modulating complement pathways in the MERTK+ macrophages could improve homeostatic function. This combined computational-experimental approach in human synovium and ex vivo systems brings insights into new complement pathway-modulated macrophage targets that upon rewiring may restore tissue homeostasis for RA. This is promising to provide a roadmap for other complex pathways in autoimmune disease tissues.
Reference
1. Zhang, F. et al. Deconstruction of rheumatoid arthritis synovium defines inflammatory subtypes. Nature 623, 616–624 (2023).
To cite this abstract in AMA style:
Vargas J, Mantel I, Inamo J, Banda N, Jonsson A, Wei K, Rao D, Goodman S, Deane K, Seifert J, Anolik J, Brenner M, Raychaudhuri S, Woodruff T, RA/SLE Network t, Holers M, Donlin L, Zhang F. Deciphering Complement-dependent Macrophage Phenotypes in Human Rheumatoid Arthritis Using Combined Computational-experimental Single-cell Omics [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/deciphering-complement-dependent-macrophage-phenotypes-in-human-rheumatoid-arthritis-using-combined-computational-experimental-single-cell-omics/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/deciphering-complement-dependent-macrophage-phenotypes-in-human-rheumatoid-arthritis-using-combined-computational-experimental-single-cell-omics/