Session Information
Date: Monday, November 18, 2024
Title: T Cell Biology & Targets in Autoimmune & Inflammatory Disease Poster
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Background Systemic lupus erythematosus (SLE) is a chronic autoimmune disease affecting multiple organs and systems. The complex pathogenesis of SLE involved the abnormal activation of CD4+T cells and DNA hypomethylation modification. Human endogenous retroviruses (HERVs) are genetic remnants of retroviruses that were integrated into the human genome millions of years ago. Transcriptional activation of HERVs may participate in the pathogenesis of autoimmune diseases.
Purpose To screen differentially expressed HERVs in CD4+T cells of SLE, explore the mechanism of regulating abnormal expression of HERVs underlying abnormal differentiation of CD4+T cells in SLE, and identify a novel target for treatment of SLE.
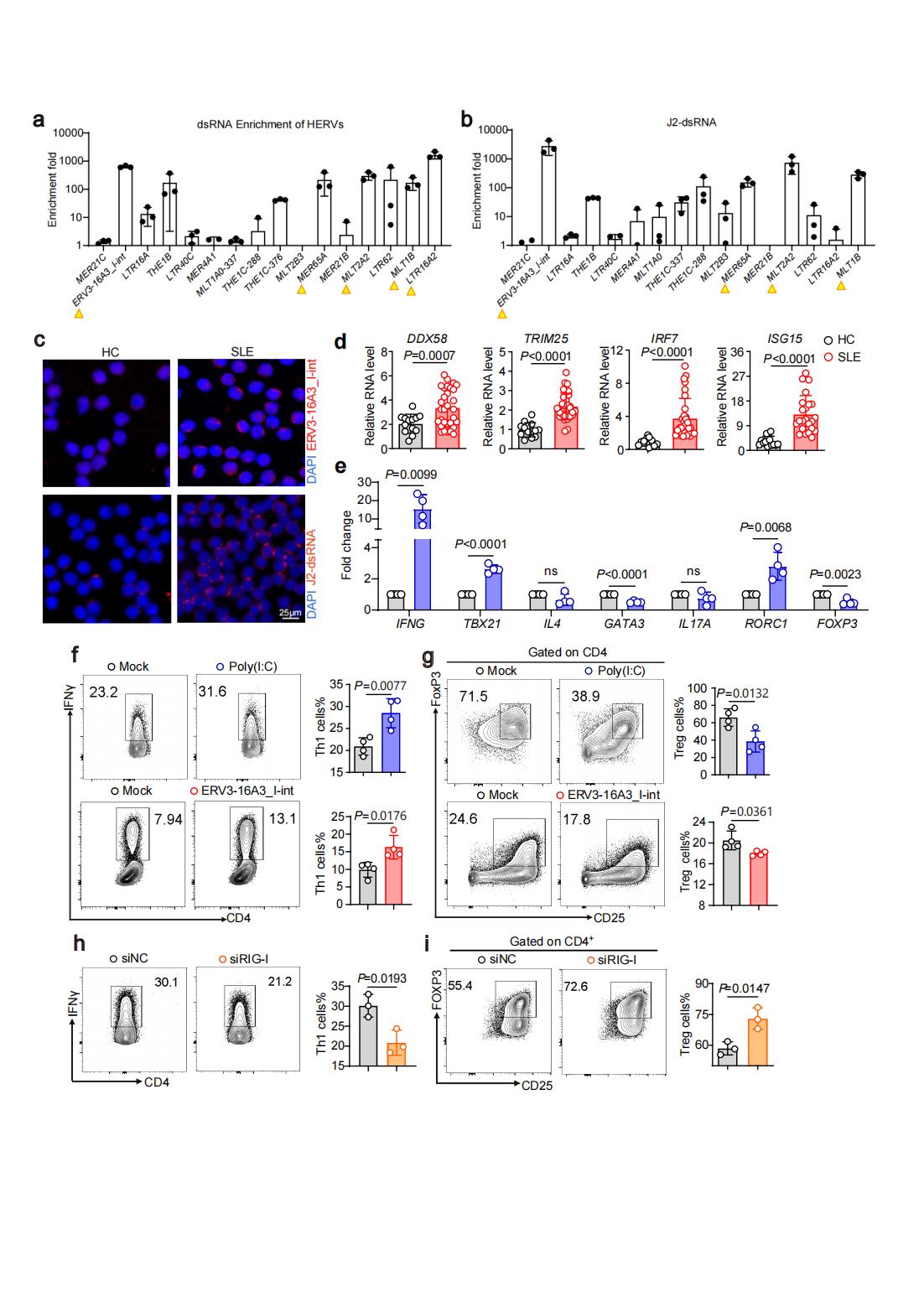
Methods: We identified HERV transcripts in CD4+T cells obtained from the whole blood of SLE patients and HCs by RNA-seq and whole transcriptome sequencing. The methylation status of these HERVs in CD4+T cells of SLE and healthy controls were investigated by Bisulfite sequencing polymerase chain reaction (BSP) assay. We performed dsRNA enrichment assay with RNase A digestion and dsRNA-specific antibody J2 pulldown assays. HERVs-derived dsRNA ERV3-16A3_I-int, poly(I:C) and silencing RNA of RIG-I were electrotransfected into human naive CD4+ T cells respectively and the cells were polarizd in vitro. The percentage of T cells was detected by flow cytometry.
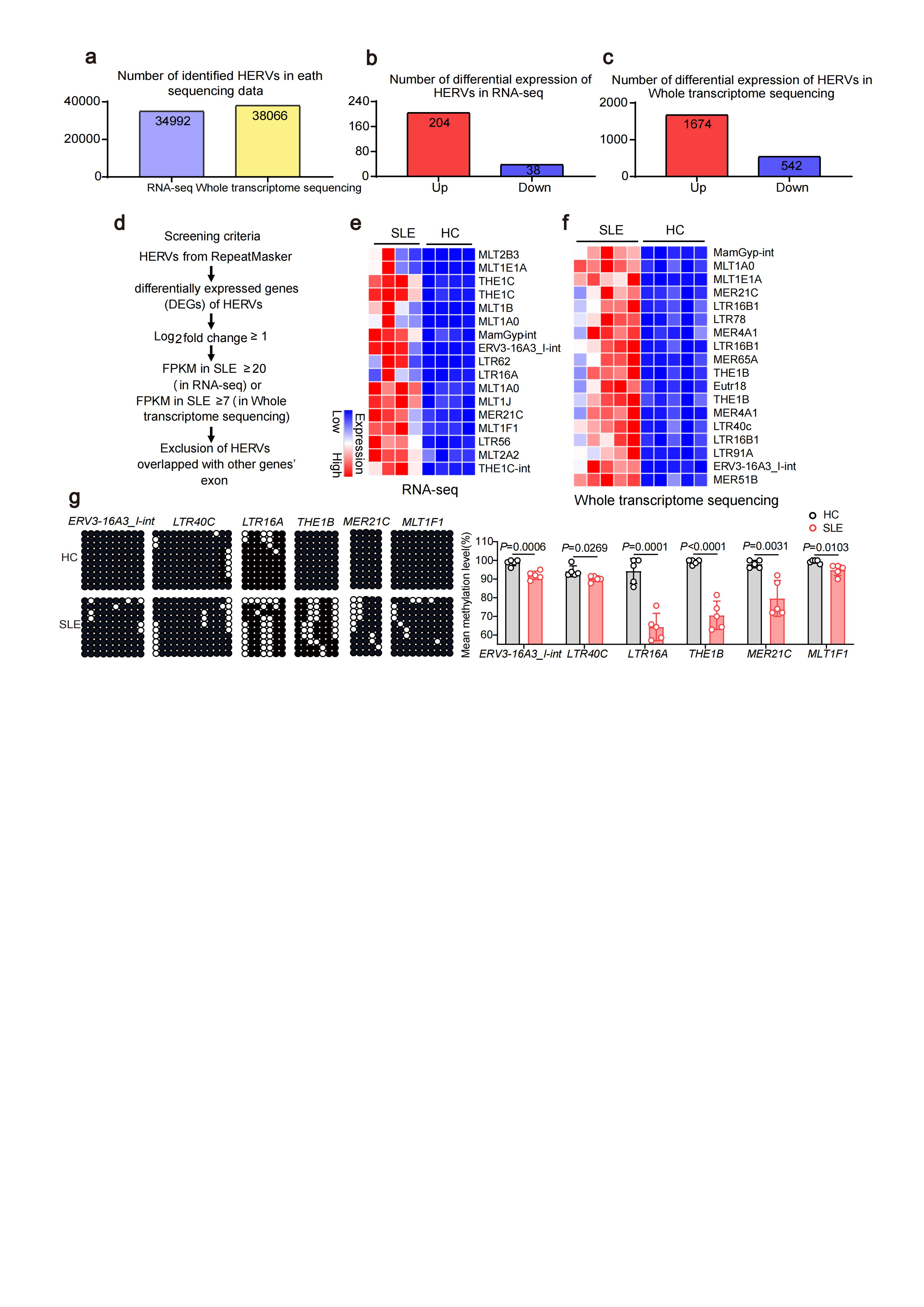
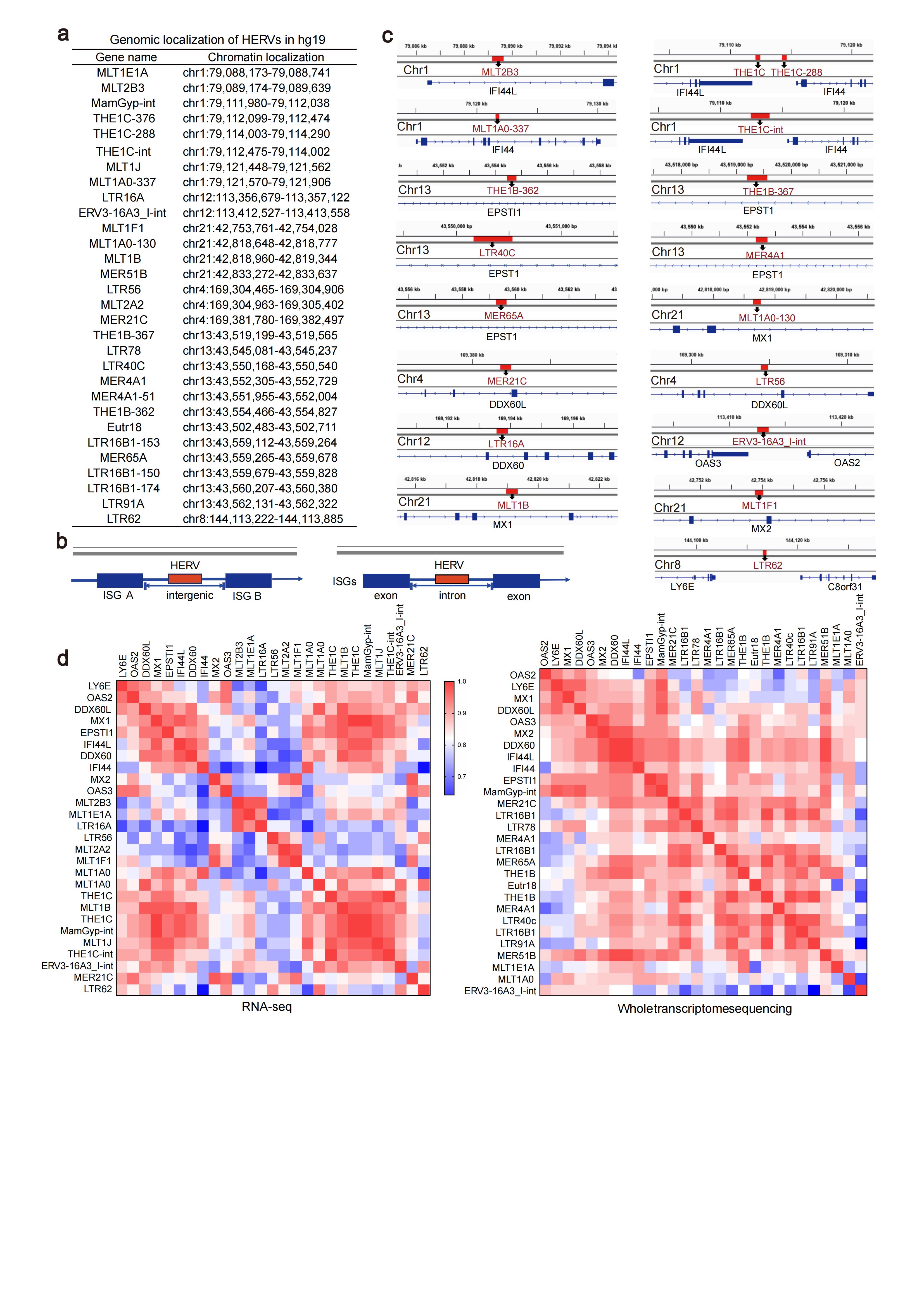
Results: We identified that many long terminal repeat sequences (LTRs), the main existence of HERVs in cells, were over-expressed in CD4+T cells of SLE (Fig.1a-f). Most of HERVs were localized in the intronic region or intergenic region of interform stimulated genes (ISGs), and the expression of ISGs was significantly positively correlated with HERVs expression (Fig.2). Moreover, we confirmed that DNA methylation status of some HERVs was shown to be significantly lower in CD4+T cells from SLE patients, and DNA methylation inhibitor promoted the transcriptional activation of HERVs, suggesting that DNA demethylation may be the cause of HERVs overexpression in SLE CD4+T cells (Fig. 1g). Among those HERVs, we found some formed double-stranded RNA (dsRNA) and the RLRs pathway in peripheral blood CD4+T cells of SLE patients were significantly up-regulated (Fig. 3a-d). Moreover, activation of RLRs signaling pathway by dsRNA (Poly(I:C) or HERVs-derived dsRNA), promoted Th1 cell differentiation and inhibited Treg cell differentiation (Fig. 3e-g). Blocking RLRs signaling pathway by silencing RIG-I inhibited Th1 cell differentiation and promoted Treg cell differentiation (Fig. 3h, i).
Conclusion: These findings suggested that DNA demethylation may be the cause of HERVs overexpression in SLE CD4+T cells, and the transcriptional activation of HERVs was closely related with pathogenic T cell differentiation in SLE. This finding uncovers the roles of HERVs and RIG-I pathway in adaptive immune response and SLE pathogenesis, and suggests that maintaining the silencing of HERVs or targeting RIG-I pathway may be a good way for SLE therapy.
To cite this abstract in AMA style:
Min X, Wu J, Yu Y, Lu Q, Zhao M. Human Endogenous Retroviruses Promote the Aberrant T Cell Differentiation in Systemic Lupus Erythematosus [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/human-endogenous-retroviruses-promote-the-aberrant-t-cell-differentiation-in-systemic-lupus-erythematosus/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/human-endogenous-retroviruses-promote-the-aberrant-t-cell-differentiation-in-systemic-lupus-erythematosus/