Session Information
Date: Sunday, November 17, 2024
Title: Abstracts: Innate Immunity: Molecular Insights Into Immune Dysregulation
Session Type: Abstract Session
Session Time: 3:00PM-4:30PM
Background/Purpose: Skin sensitivity to sunlight affects ~70% of systemic lupus erythematosus (SLE). In addition, ultraviolet radiation (UVR) can stimulate systemic disease flares, but how this occurs is unknown. To systematically examine the acute effects of UVR, we performed single cell nuclear RNA sequencing (snRNAseq) and a spatial and kinetic analysis of gene expression in the skin following UVR.
Methods: Shaved mouse skin received a single exposure of 500 mJ/cm2 UVB, and biopsies were taken at times 0 and 12 hrs (snRNAseq) and then 0, 6, and 24 hours after UVB exposure (n=3 per group) for spatial transcriptome analysis. snRNAseq employed Illumina NextSeq protocols with bioinformatics utilizing Monocle and Seurat pipelines. For spatial analysis, fixed tissue was stained with antibodies to CD45 and to cytokeratin (CK) to mark immune cells and keratinocytes, respectively and processed on the GeoMx digital spatial profiler. Mouse whole transcriptome probe set was used for next-generation sequence analysis and processed on the nanoString Software Suite 2.5.1.145. Pathway analysis was determined by Ingenuity (IPA).
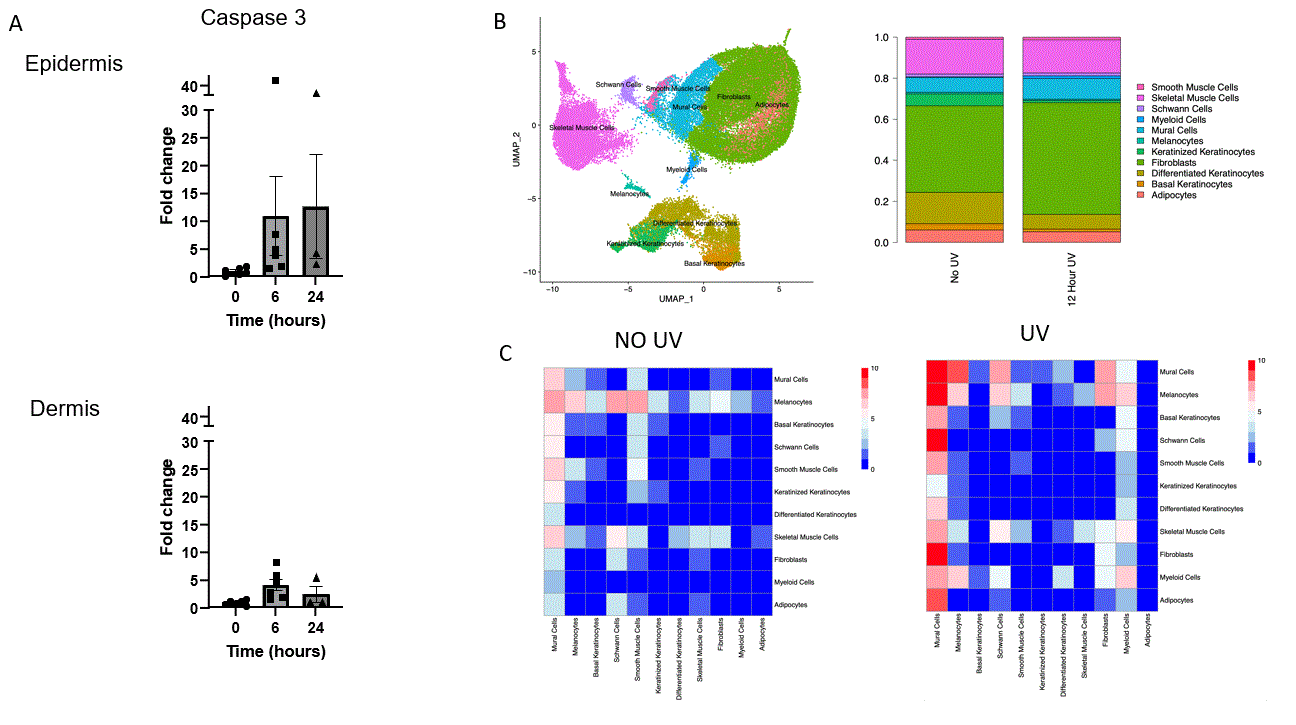
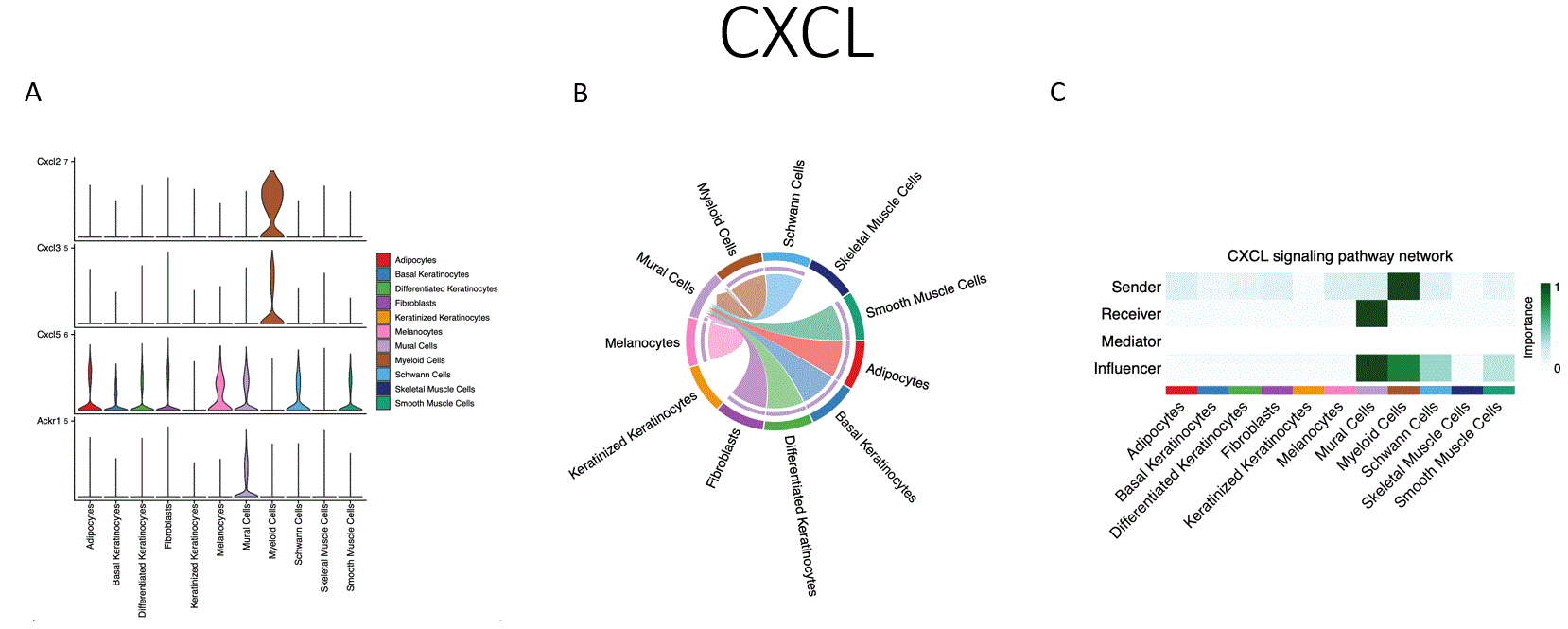
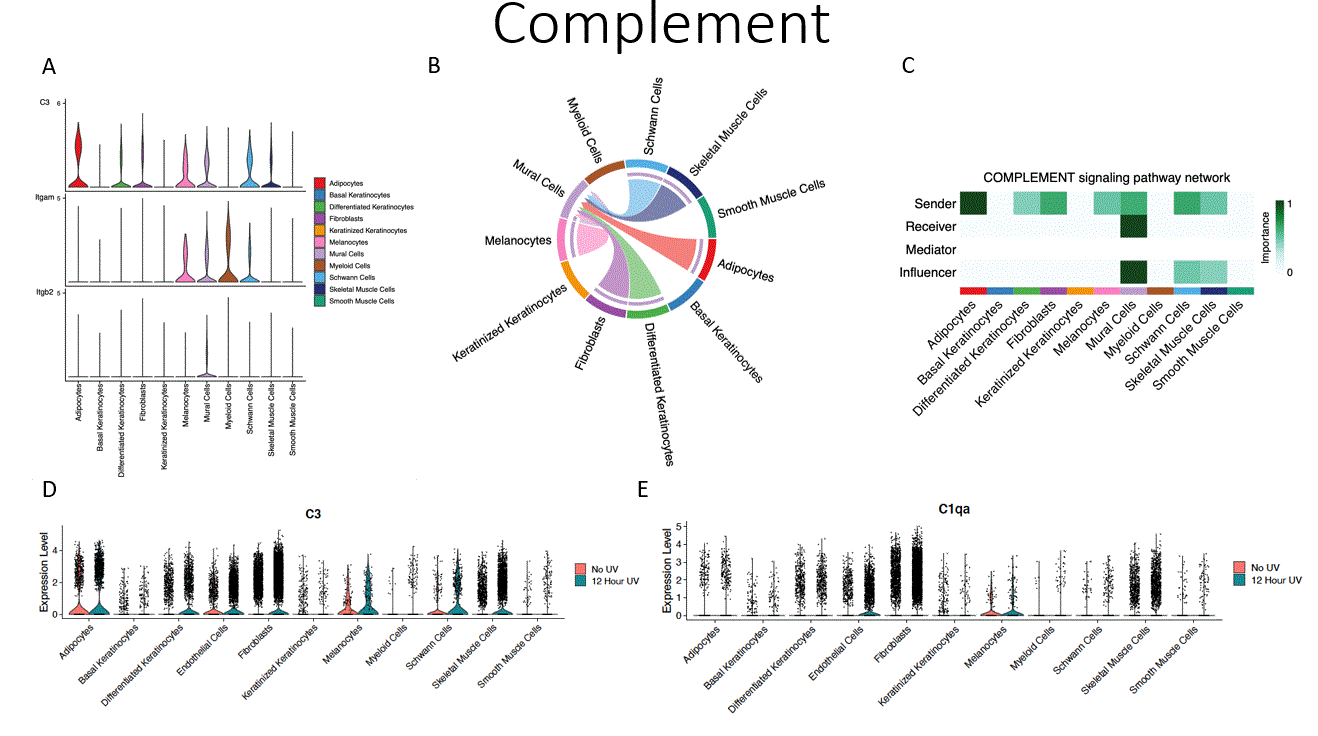
Results: UVR was accompanied by increased cellular apoptosis (caspase 3 staining), which peaked at 6 hours (Fig.1). SnRNAseq with 51,162 cells identified 12 cellular subtypes and revealed a shift toward increased myeloid and endothelial cell numbers, with decreased proportions of differentiated and keratinized keratinocytes. The most marked changes in gene expression were observed in melanocytes (MLNs), followed by endothelial cells (ECs) and keratinocytes, with lesser shifts in fibroblast and myeloid cells. Enriched biological processes shared across multiple cell types include enriched cytokine signaling in the immune system, neutrophil degranulation, senescence-associated secretory phenotype, and inflammasome pathway activation. Receptor ligand interactions demonstrated shifts towards increased activity of ECs, MLNs, and myeloid cells. Unique pathways increased with UVB exposure included CXCL chemokines (CXCL1, CXCL2, and CXCL5, Fig.2), increased CCL chemokine expression (CCL2, CCL6, CCL7, and CCL9), and complement components (C3, C4, and C1qa, Fig. 3). The major contributors to chemokine gene expression were myeloid and ECs, whereas C3 gene expression was seen across multiple cell types. These data were validated by spatial profiling (GeoMX) of the epidermis, dermis, and subcutaneous tissue. The most marked changes were in the epidermal and superficial dermal compartment with IPA highlighting apoptosis, UV-response, unfolded protein response, and p53 pathway activation and also DEGs involving leukocyte activation, cellular defense response, and regulation of reactive oxygen species.
Conclusion: Following UVR, cell stress leads to epithelial cell death and activation of inflammatory pathways. Interestingly, ECs appear to be central in orchestrating UVR inflammatory responses (Figs. 1,2,3C) that include stimulation of chemokines and complement, which in turn facilitate myeloid infiltration and immune activation. Many of these genes and pathways have been shown to be abnormal in diseases such as lupus.
To cite this abstract in AMA style:
Wasikowski R, Theisen E, An J, Tsoi L, Anufrieva K, Wei K, Gudjonsson J, Elkon K. Single Cell and Spatial Transcriptomics Implicate Vascular Cell Orchestration of Inflammatory Changes After Ultraviolet Light Exposure [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/single-cell-and-spatial-transcriptomics-implicate-vascular-cell-orchestration-of-inflammatory-changes-after-ultraviolet-light-exposure/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/single-cell-and-spatial-transcriptomics-implicate-vascular-cell-orchestration-of-inflammatory-changes-after-ultraviolet-light-exposure/