Session Information
Session Type: Abstract Session
Session Time: 3:00PM-4:30PM
Background/Purpose: Juvenile dermatomyositis (JDM) patients frequently have incomplete response to therapy. We utilized exploratory proteomics to advance understanding of dysregulated proteins and biological pathways that might contribute to disease onset, persistent disease activity and treatment response in a multi-center cohort.
Methods: We utilized a multiplexed Olink proteomics assay to measure 3072 plasma proteins in a multicenter JDM cohort, which included 56 treatment-naïve (TN) JDM enrolled at or near diagnosis (CARRA=28, U-M=7, UCSF=12, Duke=9), 24 JDM with paired six-month follow up (FU) samples (CARRA=20 and Duke=4) and 8 healthy controls (CTL). Median physician global assessment (PGA) score for TN was 6 (IQR 5-7) and for FU was 2 (IQR 1-3). We identified differentially expressed proteins (DEP) between groups, TN v CTL and FU v CTL, by fitting linear mixed effects (lmer) models using the OlinkAnalyze package (v3.5.1) in R and including “site” as a random effect. A post-hoc analysis was used to quantify the difference in protein expression between JDM and CTL. A paired t-test was used to identify DEP in paired samples (n=24) between TN and FU. Proteins associated with PGA score were identified using a lmer and individual as a random effect in CARRA samples only (n=48). Pathway analyses were performed using Ingenuity Pathway Analysis (IPA) and Genomatix Pathway System (GePS).
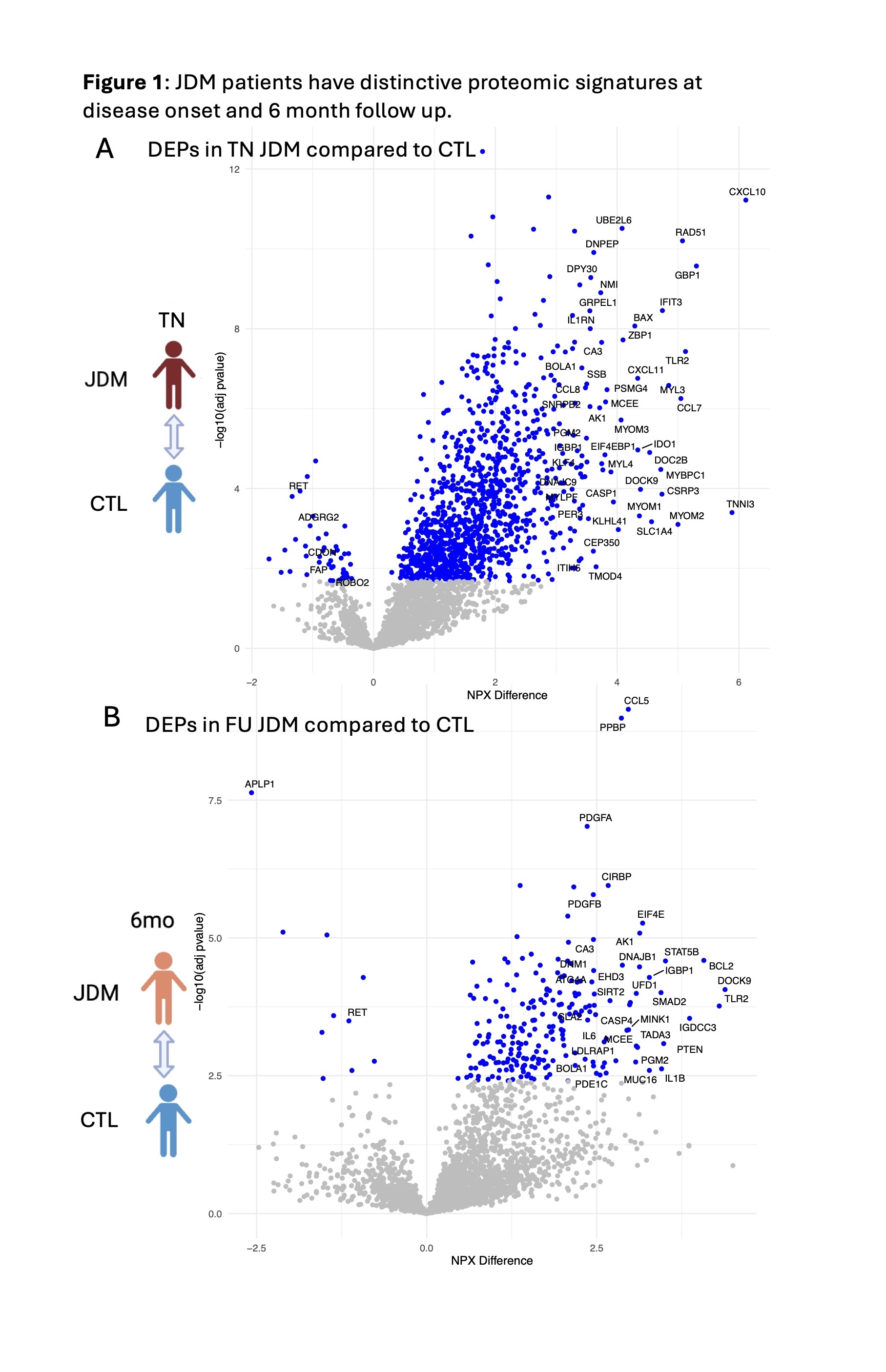
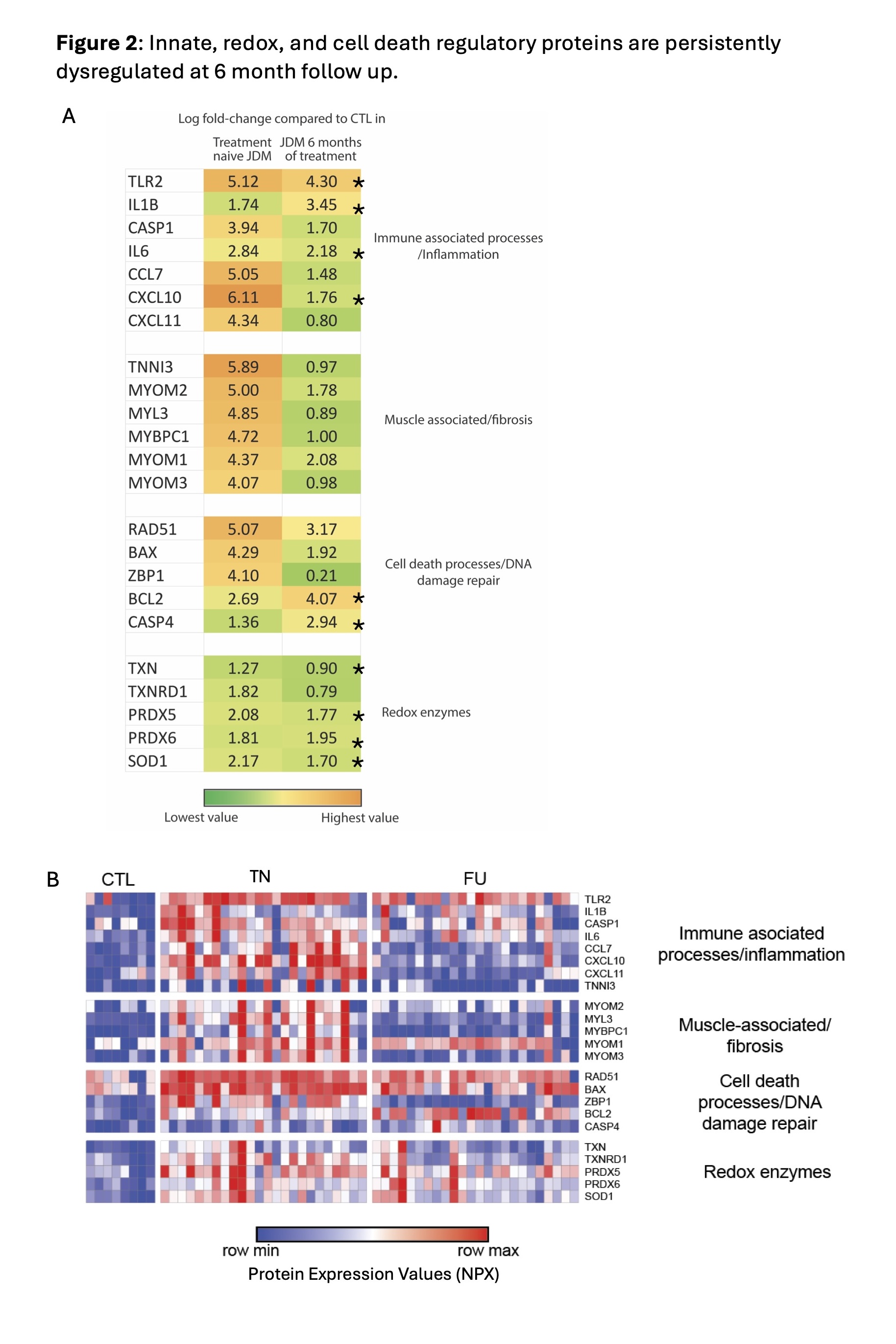
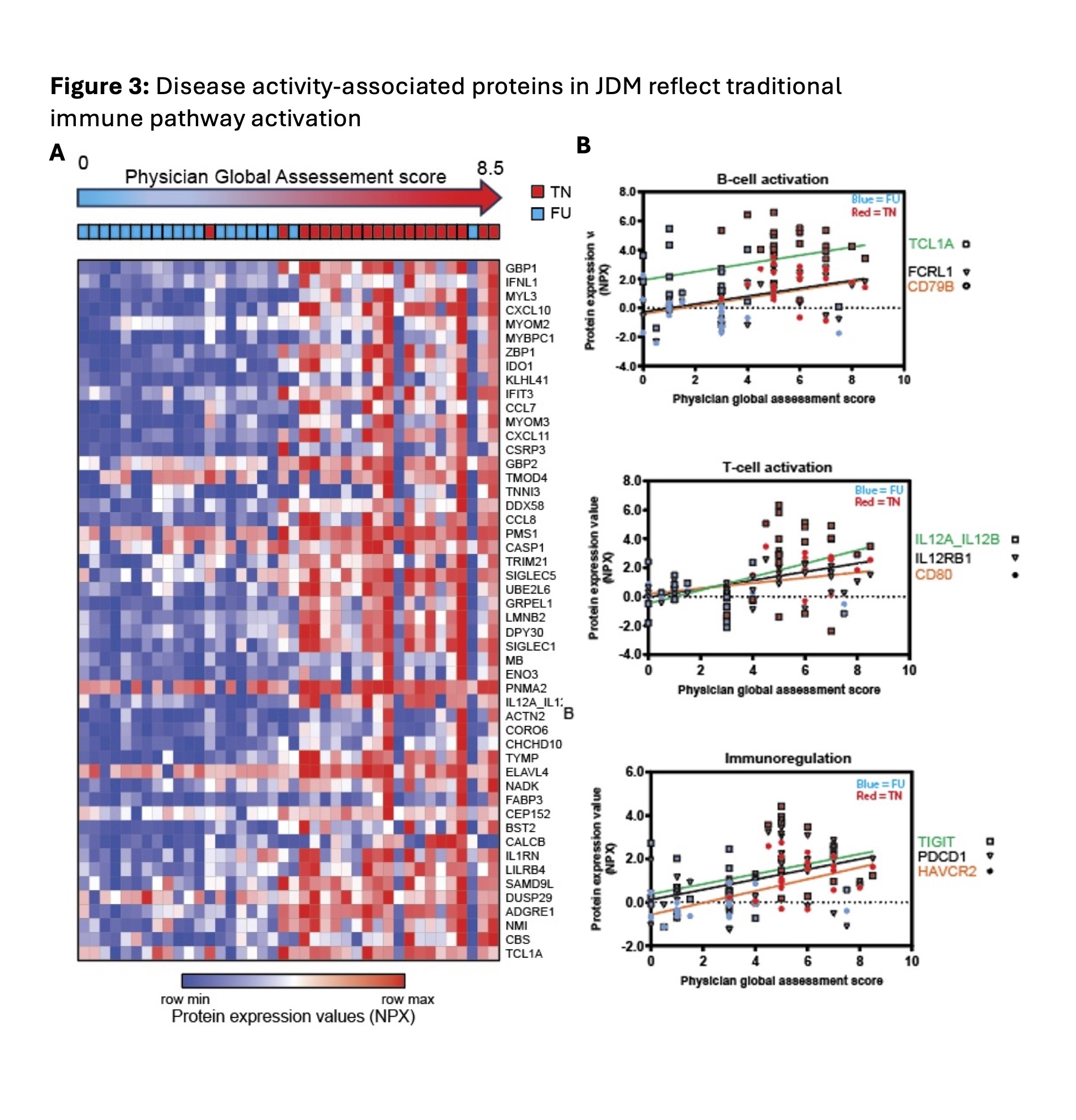
Results: There were 1176 DEP between TN JDM and CTL (Fig 1A) and 232 DEP between FU JDM and CTL (Fig 1B). We identified novel proteins differentially expressed in TN JDM enriched in regulation of cell death (BCL2, BAX, ZBP1), detoxification of ROS and response to stress (TXN, PRDX5, SOD1), muscle structure (TNNI3, MYL3, MYOM1), as well as traditional interferon-stimulated proteins (CXL10, galectin-9, SIGLEC-1) and innate immune proteins (TLR2, IL6). At FU, 196/232 of BL DEPs were still differentially expressed and included cell death regulatory, innate, and redox proteins, highlighting persistent dysregulation, with notable increases in IL1β and BCL2 (Fig 2A). A paired analysis of 24 patients with paired TN and FU samples supported these findings within individuals (Fig 2B). In 48 CARRA samples, 902 proteins (799 positively, 103 negatively) were significantly associated with global disease activity. Many top proteins were related to IFN signaling (GBP1, IFNL1, IFIT3) and muscle inflammation (MYL3, MYOM2, MYBPC1) and reflected more traditional pathways of B and T cell activation and immunoregulation (Fig 3A & B). TLR2 and RAD51 were proteins that were both persistently dysregulated at 6 month follow up as well as associated with disease activity.
Conclusion: Together, these results highlight immune pathways that are associated with JDM and decline with treatment, as well as under-recognized proteomic signatures that reflect dysregulation of cell death, detoxification of ROS, and innate immunity both at diagnosis and 6-month FU. These persistently dysregulated proteins represent potential pathways not targeted by standard induction therapy and may be more sensitive biomarkers of disease activity.
Acknowledgements: CARRA, AF, Cure JM, IRP of NIH, NIAMS
To cite this abstract in AMA style:
Neely J, Dvergsten J, Zheng Z, Madubata C, Kim H, Sabbagh S, Matossian S, Goudsmit C, Berthier C, Turnier J. JDM Proteomic Signature at Disease Onset and Progression Highlights Persistent Dysregulation of Cell Death, Redox and Innate Immune Signaling [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/jdm-proteomic-signature-at-disease-onset-and-progression-highlights-persistent-dysregulation-of-cell-death-redox-and-innate-immune-signaling/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/jdm-proteomic-signature-at-disease-onset-and-progression-highlights-persistent-dysregulation-of-cell-death-redox-and-innate-immune-signaling/