Session Information
Date: Saturday, November 16, 2024
Title: Abstracts: B Cell Biology & Targets in Autoimmune & Inflammatory Disease I
Session Type: Abstract Session
Session Time: 1:00PM-2:30PM
Background/Purpose: Anti-CD19 chimeric antigen receptor (CAR) T-cell therapy represents a promising advancement in the treatment of autoimmune diseases, including systemic sclerosis (SSc), idiopathic inflammatory myopathy (IIM), lupus nephritis (LN), and myasthenia gravis (MG) (Müller F, et al. N Engl J Med. 2024; Podoll A, et al. Lupus Science & Medicine. 2024; Haghikia A, et al. Lancet Neurol. 2023). The deep B-cell depletion possible with anti-CD19 CAR T-cell therapy may provide sustained treatment-free remission for patients with autoimmune diseases; however, a thorough understanding of the immune modulating effects remains to be elucidated. The aim of this study was to profile the cellular and molecular changes to the immune cell populations of patients with autoimmune disease before and after anti-CD19 CAR T-cell therapy.
Methods: Peripheral blood mononuclear cells (PBMCs) were collected at baseline and during early B-cell reconstitution post-CAR T-cell therapy from 6 patients with autoimmune disease, including SSc (n=2), IIM (n=2), LN (n=1), and MG (n=1), and 5 untreated healthy donors (HDs). Post-CAR T-cell sample collection coincided with clinical improvements and normalization of disease-related biomarkers. Samples were analyzed via a multi-omics approach using Verily’s Immune ProfilerTM platform. Twenty-four different subsets encompassing myeloid, T, B, and NK cells were sorted from each sample using FACS with validated cell markers. Gene expression (RNA-seq), chromatin accessibility (assay for transposase-accessible chromatin sequencing [ATAC-seq]), protein expression (targeted protein estimation by sequencing [TaPE-seq]), and next-generation sequencing were assessed. Computational analysis was performed with quality control (QC) assessment.
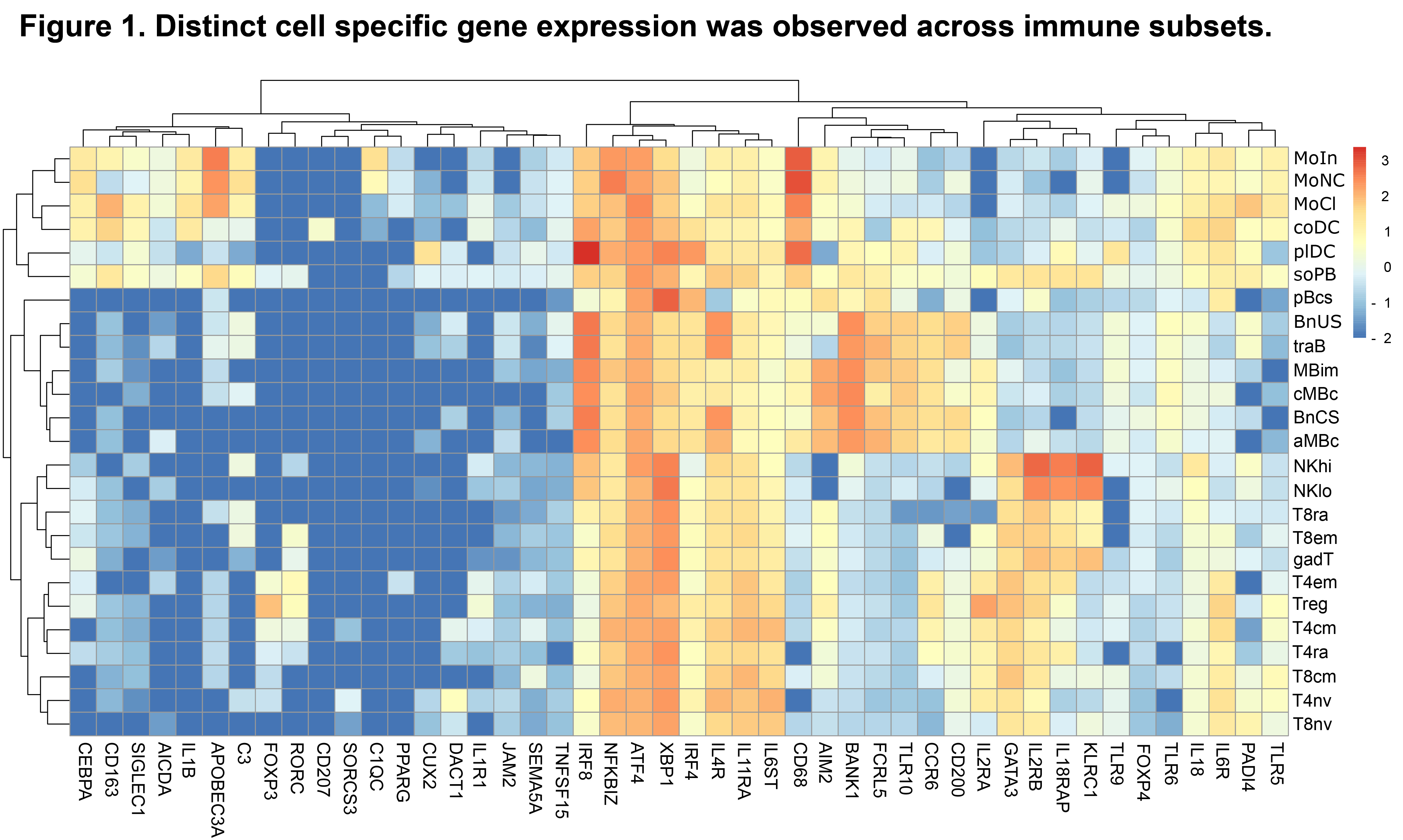
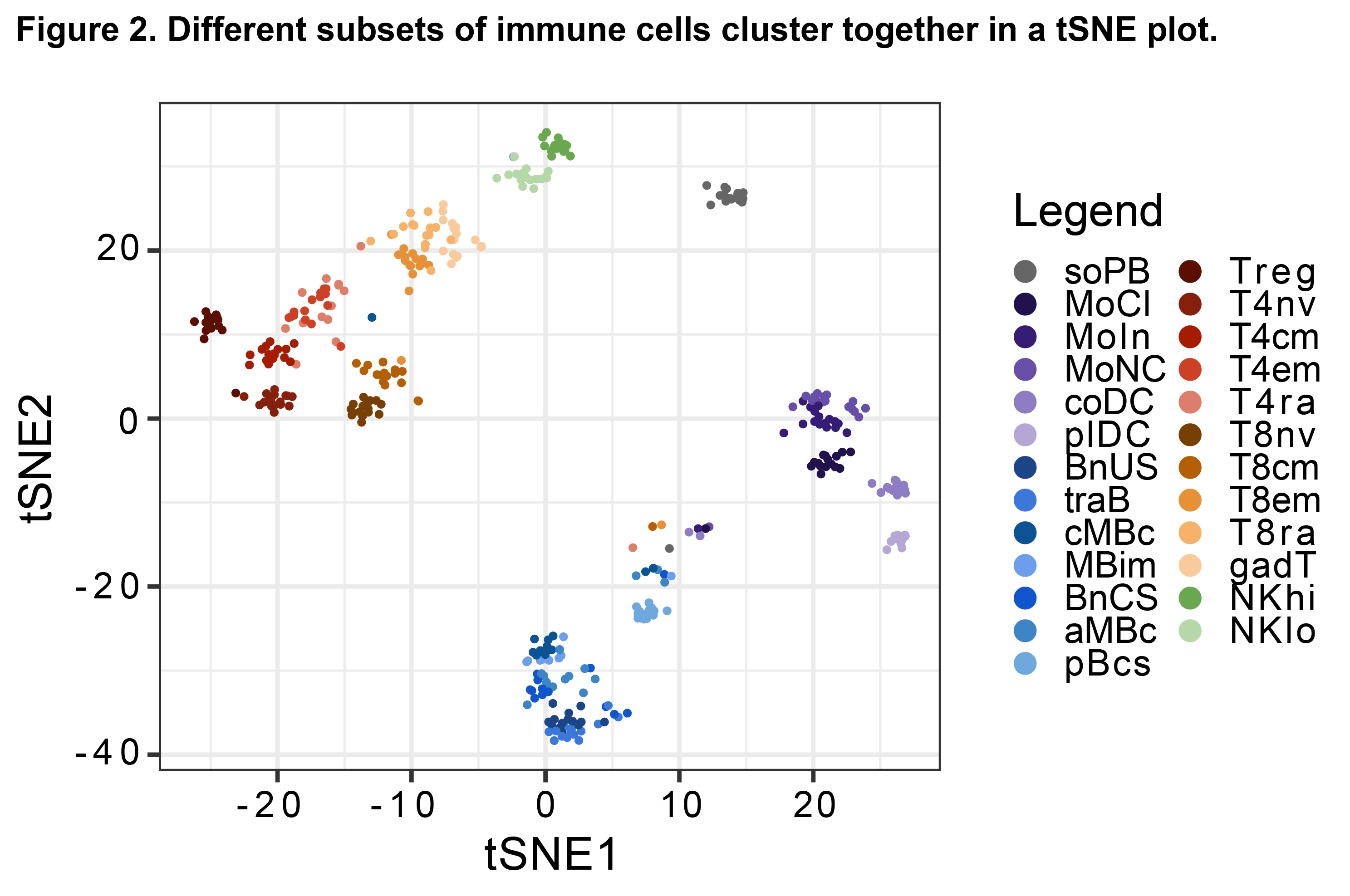
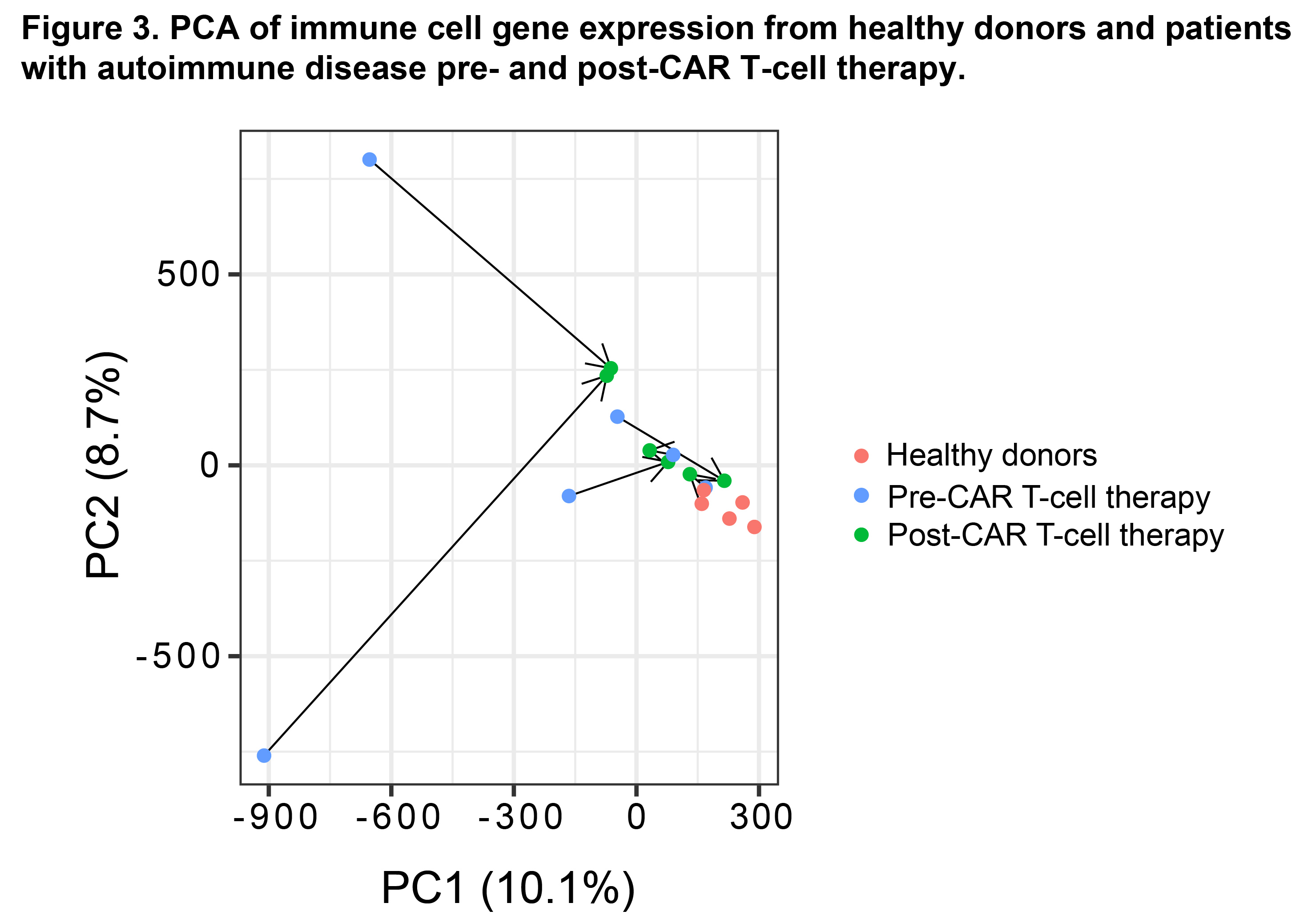
Results: A total of 1,114 sequencing datasets generated from immune cell subsets from patients and HDs met QC criteria. Immune cell type-specific gene sets were identified that aligned with published data (Fig. 1) (Ota M, et al. Cell. 2021). In addition, a t-distributed stochastic neighbor embedding plot identified distinct clusters representing each sorted immune cell subset (Fig. 2). In a pooled phenotypic analysis of all patient samples, the proportion of transitional B and CD56hi NK cells increased, and memory B and naive T cells decreased post-CAR T-cell therapy compared to baseline. A set of 6,424 differentially expressed genes, 161 differentially expressed proteins, and 53,321 differentially accessible chromatin regions (nominal p< 0.05) were observed across different immune cell subsets post-CAR T-cell therapy compared to baseline. Using principal component analysis of RNA-seq data, patients with autoimmune disease showed a trend toward convergence with HDs following CAR T-cell therapy (Fig. 3), suggestive of gene expression normalization.
Conclusion: This study demonstrates the feasibility of conducting comprehensive multi-omics analyses on PBMCs from patients with autoimmune disease pre- and post-CAR T-cell therapy. The observed molecular signatures in immune cell subsets represent promising avenues for future investigations into the mechanisms underlying the clinical effects of CAR T-cell therapy in autoimmune disease.
Abbreviations: seq, sequencing.
Abbreviations: tSNE, t-distributed stochastic neighbor embedding; seq, sequencing.
Abbreviations: CAR, chimeric antigen receptor; PCA, principal component analysis; seq, sequencing.
To cite this abstract in AMA style:
Chou J, Grieshaber-Bouyer R, Wu M, Bergmann C, Taubmann J, Müller F, Bozec A, Rothe T, MAckensen A, Podoll A, Gutman J, Haghikia A, Mougiakakos D, Tong G, Kheradpour P, Kim F, Ramesan P, Kwong B, Qu K, Ganguly B, Borie D, Chung J, Schett G. Comprehensive Immune Profiling of Anti-CD19 Chimeric Antigen Receptor T-Cell Therapy in Patients with Autoimmune Disease [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/comprehensive-immune-profiling-of-anti-cd19-chimeric-antigen-receptor-t-cell-therapy-in-patients-with-autoimmune-disease/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/comprehensive-immune-profiling-of-anti-cd19-chimeric-antigen-receptor-t-cell-therapy-in-patients-with-autoimmune-disease/