Session Information
Date: Saturday, November 16, 2024
Title: Muscle Biology, Myositis & Myopathies – Basic & Clinical Science Poster I
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: Sporadic inclusion body myositis (sIBM) is often challenging to diagnose because many patients have no known biomarkers (seronegative). Machine learning was applied to identify novel sIBM autoantibodies on a human proteome microarray (HuProt) and proteins in sera and muscle biopsies using mass spectrometry (MS)-based quantitative shotgun proteomics.
Methods: Sera from sIBM patients (n=10) were screened for autoantibodies using HuProtTM array (PEPperPRINT, Germany). Sera (n=5) and muscle biopsy tissues (n=10) from sIBM patients were screened for proteins using MS. Traditional log-fold change method and machine learning feature selection techniques (permutation importance, select K best, recursive feature elimination) were used to select the most important biomarkers for differentiating between sIBM and healthy controls. We examined the potential function of selected biomarkers (absolute log-fold change threshold >2, p < 0.05) in potential pathways of sIBM pathogenesis (autoimmune, inflammation, and degenerative pathways) using Genome Ontology (GO) analysis.
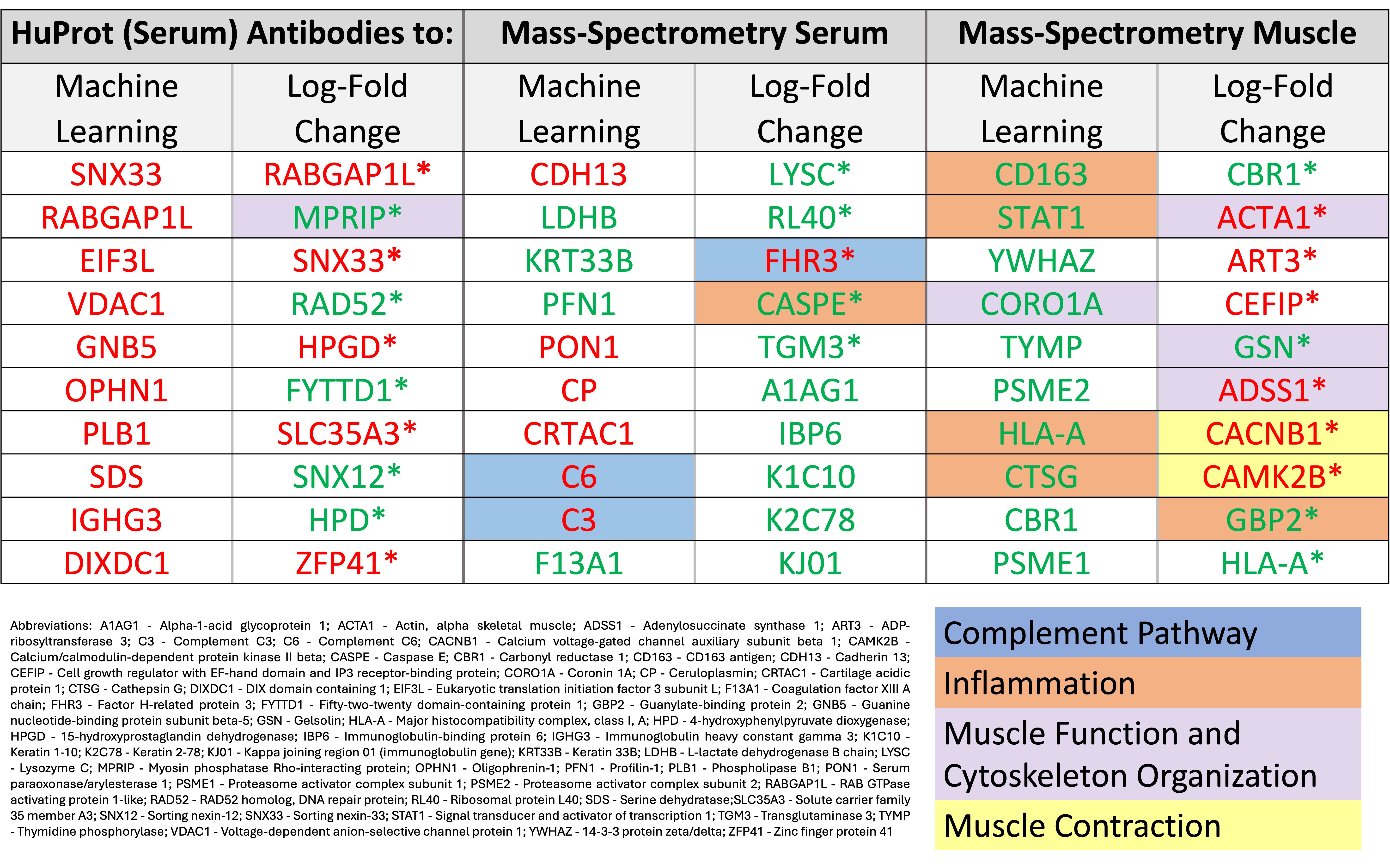
Results: 12,319 autoantibodies were detected using HuProt and 438 proteins were detected in sera and 2462 proteins in muscle biopsies using MS. Traditional log-fold change method and/or machine learning feature selection identified immune-mediated biomarkers involved in 1) complement activation (decreased C3, C6, complement factor H related 3 expression) and 2) inflammation (increased CD163, STAT1, HLA-A, cathepsin G, caspase 14, and guanylate binding protein 2 (GBP2)) (Table). Related to muscle function, there were biomarkers involved in 1) cytoskeleton organization (increased coronin 1A (CORO1A), gelsolin (GSN), and anti-myosin phosphatase Rho interacting protein (MPRIP) antibodies, as well as decreased actin alpha 1 (ACTA1), adenylosuccinate synthase 1 (ADSS1, mutation causes distal myopathy)) and 2) muscle contraction (decreased calcium voltage-gated channel auxiliary subunit beta 1 (CACNB1) and calcium/calmodulin dependent protein kinase II beta (CAMK2B)).
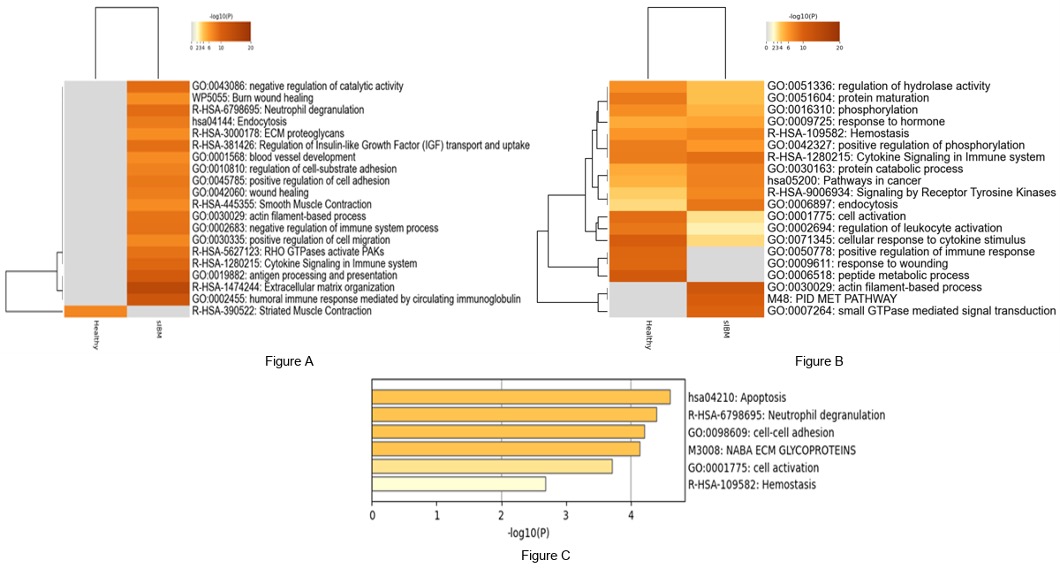
GO confirmed that apoptosis, neutrophil degranulation, cytokine signaling, antigen processing and presentation, and humoral immunity were upregulated in sIBM compared to controls (Figure). There were several pathways related to muscle function involved in sIBM including a downregulation of striated muscle contraction and upregulation of smooth muscle contractions and actin-filament based processes compared to controls.
Conclusion: Several novel sIBM biomarkers involved in the immune and degenerative pathways were identified using proteome microarray and MS proteomics with the aid of machine learning analysis. These biomarkers may improve sIBM diagnosis and our understanding of disease pathogenesis. Future studies validating our findings in a larger cohort of patients are needed prior to the development of a diagnostic platform for clinical use.
To cite this abstract in AMA style:
Moghaddam F, Tarnopolsky M, Leclair V, Hudson m, Mitchell R, Buhler K, Hatcher E, Zhang M, Dufour A, de Almeida L, Fritzler M, Choi M. Sporadic Inclusion Body Myositis Novel Autoantibody and Biomarker Research Utilizing Proteome Microarray and Mass Spectrometry Proteomics Analysis [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/sporadic-inclusion-body-myositis-novel-autoantibody-and-biomarker-research-utilizing-proteome-microarray-and-mass-spectrometry-proteomics-analysis/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/sporadic-inclusion-body-myositis-novel-autoantibody-and-biomarker-research-utilizing-proteome-microarray-and-mass-spectrometry-proteomics-analysis/