Session Information
Date: Tuesday, November 14, 2023
Title: Abstracts: Muscle Biology, Myositis & Myopathies – Basic & Clinical Science I
Session Type: Abstract Session
Session Time: 2:00PM-3:30PM
Background/Purpose: Autoantibodies directed against various tRNA synthetases defines the anti-tRNA synthetase syndrome (ARS). While it is unclear if the autoantibodies are directly pathogenic, the efficacy of rituximab in ARS implies that B cell dysregulation is critical in disease pathogenesis. We investigate how B cell transcriptomics varies in patients with active ARS versus healthy controls (HCs).
Methods: Ten patients with active ARS and HCs were identified from the MYSTIC cohort (VUMC IRB 141415). We thawed cryopreserved peripheral blood mononuclear cells (PBMCs) and isolated live CD19+ B cells using fluorescence activated cytometry sorting. Single cell RNA-Seq was performed using the 10X chromium platform with a target depth of 50,000 reads/cell. Data were de-multiplexed and processed using the CellRanger pipeline, and single-cell analysis was performed using Seurat v4.0.0. Immunoglobulin, MHC, and Y-chromosome genes were removed to prevent them driving transcriptionally defined clusters. B cell subset identities were assigned to clusters based on prior literature transcriptional profiles. Differential gene expression analysis comparing was performed in Seurat, subjected to GSEA pathway analysis, and visualized with CytoScape.
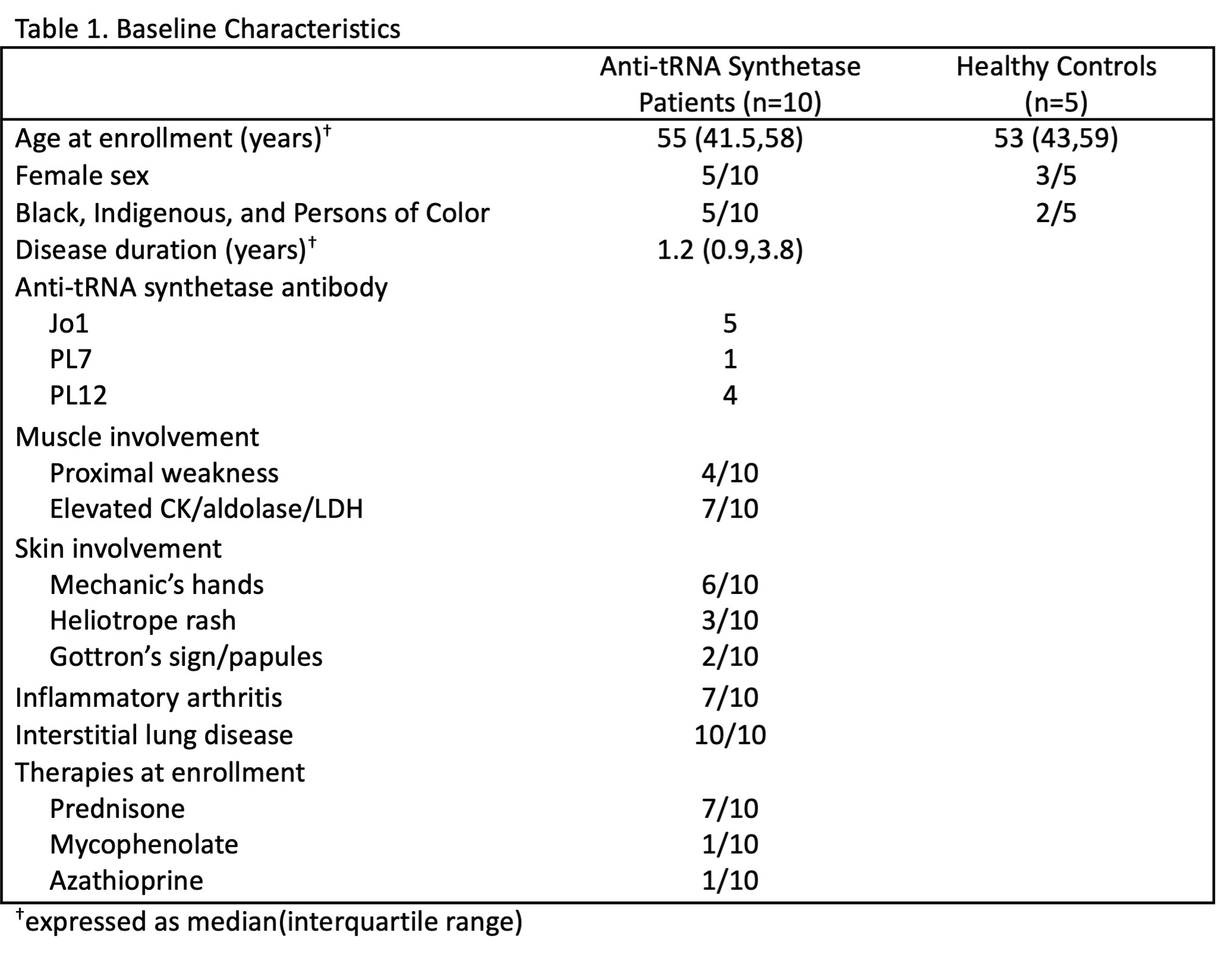
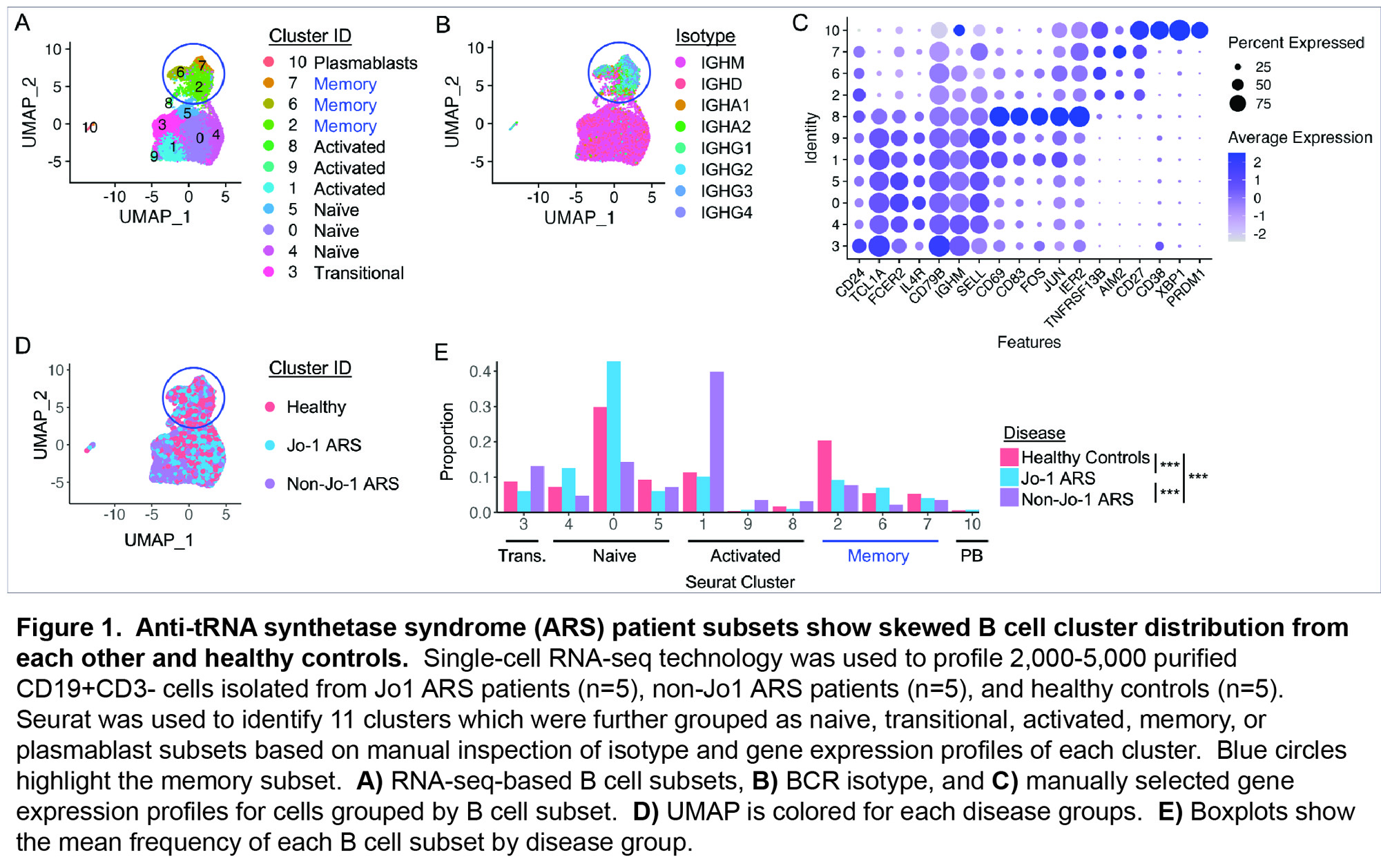
Results: Table 1 shows baseline participant characteristics. Eleven unique transcriptional clusters were identified and collapsed into five B cell subsets (transitional, naïve, activated, memory, plasmablast (Fig. 1A)) for downstream analysis based on isotype switching and gene expression (Fig. 1B-C). Initial visual examination of Jo-1 ARS, non-Jo-1 ARS, and healthy control groups on UMAP plots suggested potential B cell subset skewing across participant groups (Fig. 1D). Jo-1 ARS patients showed an increase in naïve B cells, Non-Jo-1-ARS patients had increased activated B cells, and both ARS patient groups tended to have a lower frequency of memory B cells than HCs (Fig. 1F). Results of the GSEA analysis are shown in Fig. 2. Notably, the gene ontology term “defense response to symbiont,” which contains the interferon genes IFI6, IFI44L, IFITM1, IFITM2, IRF7, ISG20, MX2, STAT1, was the most overrepresented term for transitional (FDR p=0.009) and activated (FDR p< 0.001) subsets. The Reactome term “Eph-ephrin signaling,” which contains the critical B cell activation/migration genes ACTB, AP2M1, CDC42, CFL1, RAC1, and RHOA, was the most overrepresented term for memory cells (FDR p=0.01).
Conclusion: B cell subset frequencies are skewed in ARS compared to HCs. ARS patients have altered B cell and interferon signaling compared to healthy controls. Future work will dissect how ARS B cell activation differs from normal immune responses.
To cite this abstract in AMA style:
Wilfong E, Crofford L, Bonami R. B Cells in Anti-tRNA Synthetase Syndrome Patients Show an Activated, Interferon Responsive Signature [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/b-cells-in-anti-trna-synthetase-syndrome-patients-show-an-activated-interferon-responsive-signature/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/b-cells-in-anti-trna-synthetase-syndrome-patients-show-an-activated-interferon-responsive-signature/