Session Information
Date: Tuesday, November 14, 2023
Title: (1776–1795) Spondyloarthritis Including Psoriatic Arthritis – Basic Science Poster
Session Type: Poster Session C
Session Time: 9:00AM-11:00AM
Background/Purpose: Psoriatic arthritis (PsA) is a form of inflammatory arthritis that occurs in patients with cutaneous psoriasis. Due to the lack of clinically useful diagnostic biomarkers or full understanding of pathways distinguishing PsA from psoriasis without PsA (PsC), we aimed to conduct integrated multiomics analyses using data from previously identified panel of SNPs and proteins, peripheral blood bulk RNA sequencing, miRNA sequencing and serum metabolomics obtained from carefully phenotyped individuals with psoriatic disease to identify pathways that differentiate PsA from PsC.
Methods: Serum, RNA and DNA from 102 PsA and 100 PsC patients were retrieved from a biobank of patients with psoriatic disease. Serum samples were used for miRNA sequencing, ELISA of 15 proteins (TNFSF14, S100A8/9, COMP, CRP, M2BP, OPG, DEFA, ITGB5, RANKL, CXCL10, Leptin, MMP3, OPN, Periostin, and SOST) and liquid chromatography-high resolution mass spectrometry (LC-HRMS) for metabolites, RNA for RNAseq and DNA for genotyping 42 SNPs of 19 ‘PsA weighted’ genes. Resulting molecular data were combined with clinical data to identify the best set of biomarkers able to discriminate between PsA and PsC. A random forest model was developed to identify the best combination of diagnostic biomarkers. Further bioinformatics analysis was conducted using mirDIP 4.11 with threshold “very high” to identify biomarker mRNAs targeted by biomarker miRNA, and pathDIP 42 was used to perform pathway enrichment analysis for such mRNAs, while clusterProfiler 4.8.03 was used to perform Gene Ontology enrichment analysis. Analyses and visualization were performed in R 4.3.0.
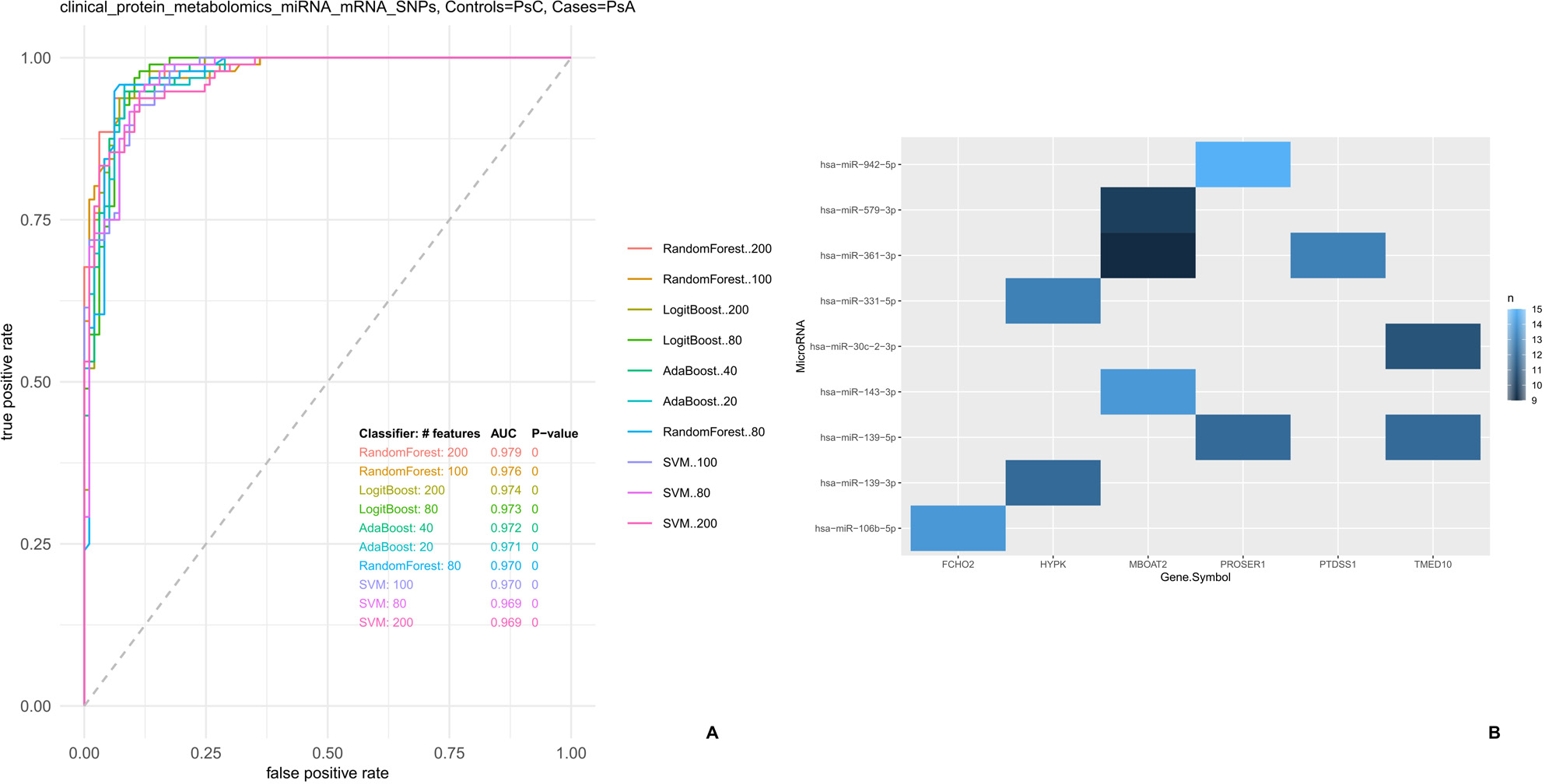
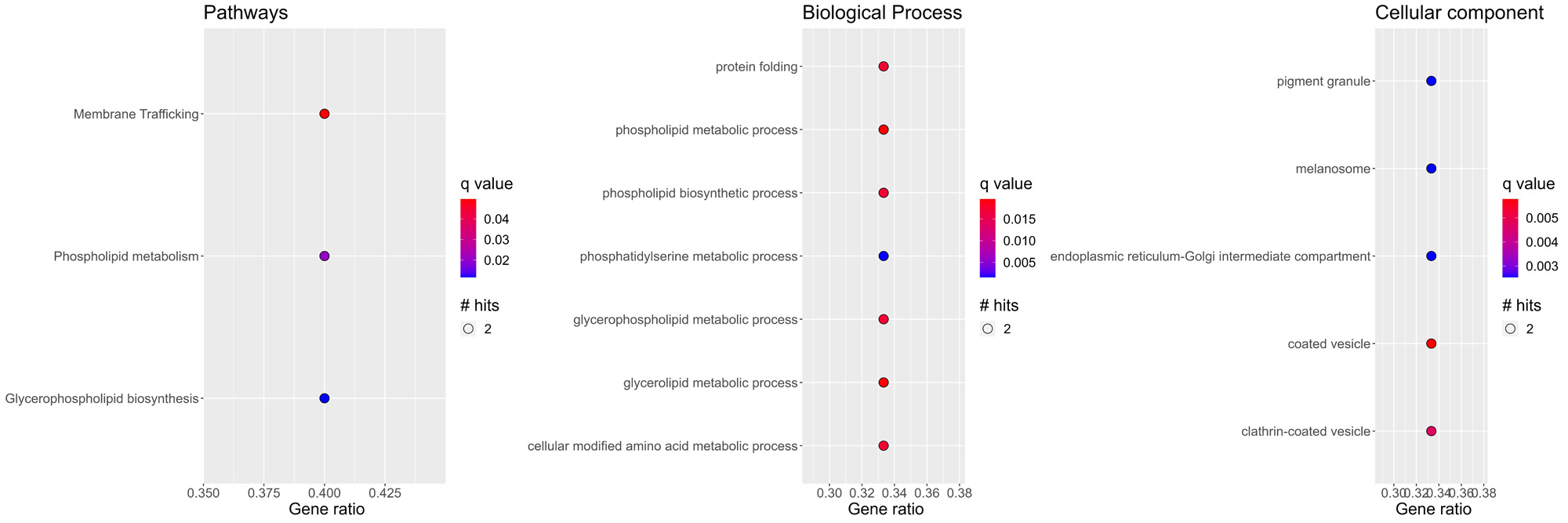
Results: The random forest model identified 200 biomarkers, comprising 97 metabolites, 77 miRNAs, and 26 mRNAs, as the best performers to discriminate between PsA and PsC (Figure 1A, AUROC of 0.979, p-value < 0.001).Among the 77 miRNAs, 9 target 6 of the 26 mRNAs present in the 200 biomarkers (Figure 1B).Pathway and Gene Ontology enrichment analyses performed on the 6 genes identified several terms linked to phospho- and glycerophospholipid metabolism (Figure 2).Interestingly, of the 97 biomarker metabolites, 40 were mapped to known metabolites. Of these, 12 (30%) were either glycero- or phospholipids. Differences in phospholipid profiles between PsA and PsC have been described in 4, where a link to the immune system has also been proposed. Further validation is undergoing to confirm the diagnostic value of the 200 biomarkers and the role of the phospholipidic pathways in PsC progression to PsA.
Conclusion: Our study highlights the importance of integrating multiple types of data to characterize different and complementary pieces of the molecular landscape that differentiate PsA and PsC. Our preliminary analysis shows that metabolic pathways can have a central role in the development and differentiation of PsA, a signal identified at the genetic level and confirmed through metabolomics analysis.
References: 1. 1.Tokar, T. et al.Nucleic Acids Res46, D360 (2018). 2. 2.Rahmati, S. et al.Nucleic Acids Res48, D479–D488 (2019). 3. 3. Wu, T. et al.Innovation (Cambridge (Mass.))2, 100141 (2021). 4. 4.Wójcik, P. et al.Int J Mol Sci20, (2019).
To cite this abstract in AMA style:
Pastrello C, Ganatra D, Kotlyar M, Looby N, Li Q, Eder L, Gladman D, Rahman P, Jurisica I, Chandran V. Multi-Omics Analyses Identify Metabolic Pathways That Differentiate Psoriatic Arthritis from Psoriasis [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/multi-omics-analyses-identify-metabolic-pathways-that-differentiate-psoriatic-arthritis-from-psoriasis/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/multi-omics-analyses-identify-metabolic-pathways-that-differentiate-psoriatic-arthritis-from-psoriasis/