Session Information
Date: Monday, November 14, 2022
Title: Abstracts: SLE – Diagnosis, Manifestations, and Outcomes III: Genetic Factors
Session Type: Abstract Session
Session Time: 3:00PM-4:30PM
Background/Purpose: SLE is an inflammatory condition associated with hyperactivation of the immune system, with mounting evidence that imbalances in the gut microbiota communities are common. These imbalances can range from subtle patterns of dysbiosis to blooms in abundance of individual species that are concordant with clinical disease flares. Based on preliminary longitudinal surveys, more than 40% of Lupus nephritis (LN) flares were concurrent with transient expansions of a pathobiont, Ruminococcus (blautia) gnavus. As the transcriptomic patterns in the cells in our bloodstream can reflect disease activity, we sought to investigate gene expression patterns in groups of lupus patients, with comparisons to healthy controls (HC).
Methods: From a well-characterized cohort, exploratory studies were performed on a selected group of 15 active female SLE patients, based in part on SLEDAI scores > 4. Patients were grouped as without a history of renal involvement (i.e., non-renal) (N=7) or with LN in flare with Urine Protein creatinine ratio >0.5. Based on 16S rRNA fecal microbiota analyses, LN were subsetted as without bloom of individual species (N=4), or with a bloom ( > 20-fold increased from HC, 3-9% abundance) of R. gnavus (N=4). In comparisons with 8 female HC, bulk RNA-seq was completed and after standard QC and filtering, 24,319 genes passed a minimum threshold of 4 reads in >50% samples and were used in downstream analysis.
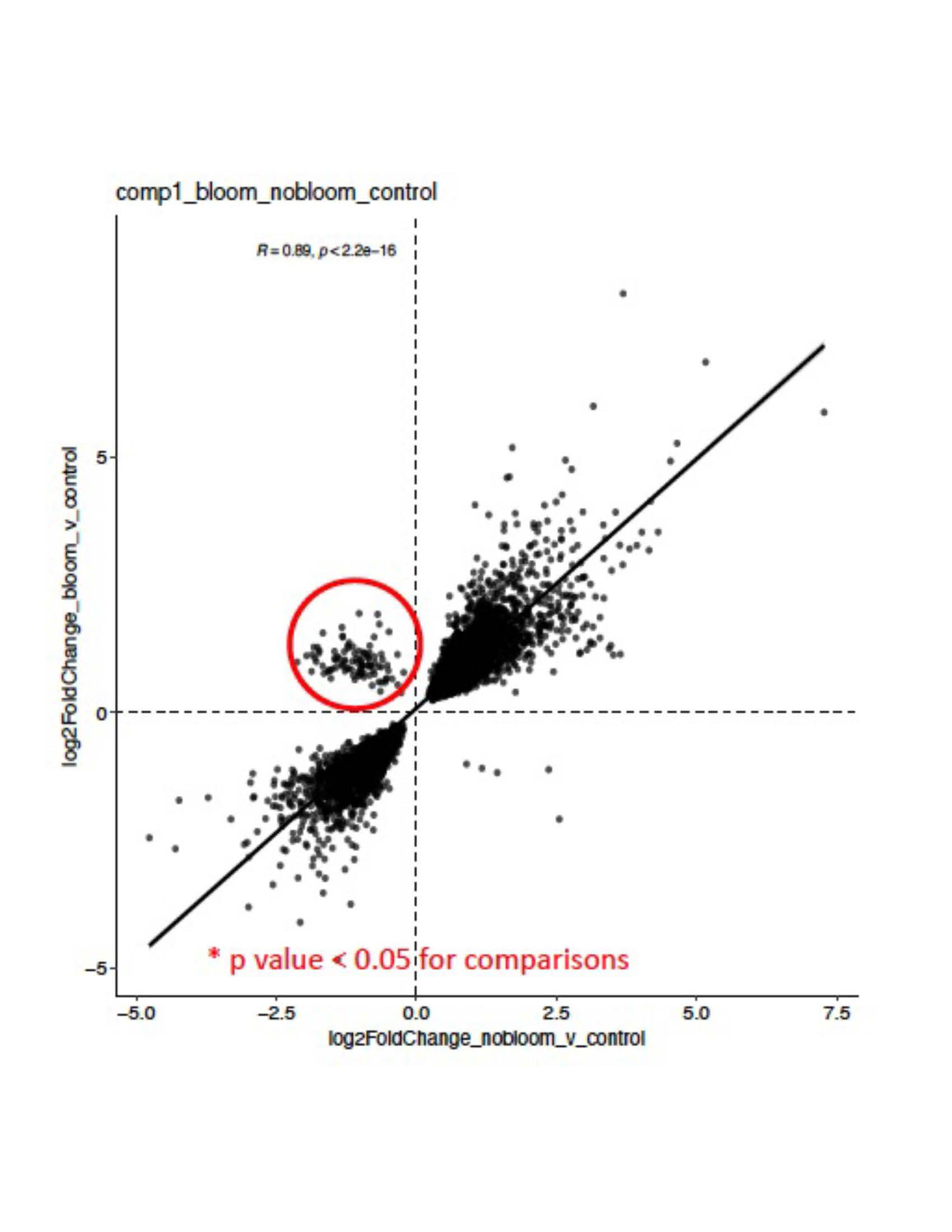
Results: Unsupervised clustering based on gene expression demonstrated significant separation between SLE samples and HC (Figure 1). While there was no clear distinction between non-renal and renal (i.e., LN) groups, there were striking differences in the group of active LN without detectable gut microbiome blooms vs. active LN with R. gnavus blooms (Figure 2), with 173 upregulated and 13 downregulated genes based on differential expression analysis (padj < 0.05, log2fc > 1). Gene set enrichment analysis (GSEA) identified several significantly altered pathways in the LN flare with blooms compared to no blooms, in highlighting multiple platelet activation pathways in the LN with blooms. In contrast, the LN flares without blooms had significantly higher interferon alpha and interferon gamma signatures.
Conclusion: Our findings document distinct pathways of immune activation in PBMC of LN in flare with concurrent gut blooms of R. gnavus, a commensal that is well-established as expanded in active LN patients, including an independent SLE cohort studied at initial diagnosis without previous treatment. These findings may indicate that in a major subset of patients, LN flares may arise from severe perturbations of intestinal communities associated with a leaky gut, such as R. gnavus. Moreover, R. gnavus gut colonization has been shown in in vivo models to stoke a feed-forward pro-inflammatory response associated with raised autoantibody and anti-bacterial antibody responses. Together, this highlights R. gnavus as a potential causative agent for Lupus flares, in which blooms may induce immune hyperactivation that underlies lupus autoimmune pathogenesis.
To cite this abstract in AMA style:
Silverman G, Cornwell M, Izmirly P, Masson M, Buyon J, Azzouz D, Ruggles K. Lupus Clinical Flares in Patients with Gut Pathobiont Blooms Share a Novel Peripheral Blood Transcriptomic Immune Activation Profile [abstract]. Arthritis Rheumatol. 2022; 74 (suppl 9). https://acrabstracts.org/abstract/lupus-clinical-flares-in-patients-with-gut-pathobiont-blooms-share-a-novel-peripheral-blood-transcriptomic-immune-activation-profile/. Accessed .« Back to ACR Convergence 2022
ACR Meeting Abstracts - https://acrabstracts.org/abstract/lupus-clinical-flares-in-patients-with-gut-pathobiont-blooms-share-a-novel-peripheral-blood-transcriptomic-immune-activation-profile/