Session Information
Date: Monday, November 9, 2020
Title: Spondyloarthritis Including Psoriatic Arthritis – Basic Science Poster
Session Type: Poster Session D
Session Time: 9:00AM-11:00AM
Background/Purpose: Perturbations of the gut microbiota have been associated with Psoriatic Arthritis (PsA), a chronic inflammatory disease. We aim to test the microbiome-metabolic interface of patients with PsA when compared to healthy controls (HC), this phenomic approach study the microorganism composition of the gut and specific gut-microbial-related metabolites
Methods: This was an observational study conducted in two centres at the United Kingdom. Stool, urine and serum samples from 19 PsA patients and 12 HC were collected. Stool microbial profiles were generated using 16S rRNA gene sequencing (using Illumina MiSeq and Oxford Nanopore Sequence). Fecal, serum and urine supernatants were analysed using a Bruker Avance IIII 600 MHz spectrometer. In-house scripts were used to process NMR spectra (Figure 1).
Results: Quantification of small molecules in serum samples; Tyrosine (p-value < 0.05), Phenylalanine (p-value < 0.05), Histidine (p-value < 0.05) and Alanine (p-value < 0.005) were increased in PsA participants and Glutamine (p< 0.005) was increased in the HC (Figure 2). The 16s rRNA gene sequencing reported an increase in the relative abundance of the family, Lachnospiraceae (p-value < 0.005) and one of its genus, Blautia (p-value < 0.005), taxa that have been positively associated with serum alanine levels in the literature (Figure 3).
Conclusion: While there are confounders in this pilot study which may contribute to findings, we found significant changes in the amino acids related to the gut microbiome in PsA patients. New associations were made for PsA when compare with previous studies of chronic inflammatory diseases. Further work will validate these results and disclose further links between PsA and the gut microbiota.
 Serum multivariate analysis techniques (A) Serum – PCA (B) Serum O-PLS after 1000 permutations (p-value = 0.0250)
Serum multivariate analysis techniques (A) Serum – PCA (B) Serum O-PLS after 1000 permutations (p-value = 0.0250)
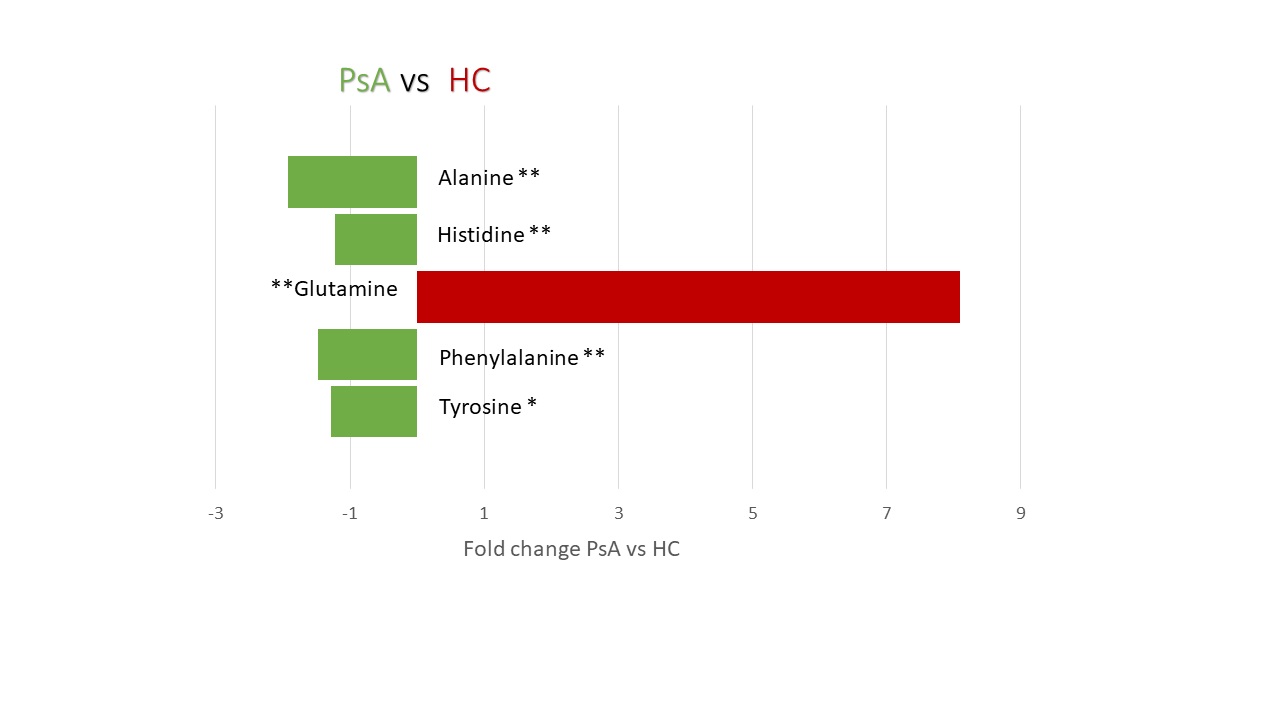 Changes in the two groups of participants in the study (19 PsA ; 12 HC ). * p-value < 0.005 ** p-value < 0.005. Quantification of small molecules was performed with a simplex approach based algorithm developed in-house and implemented in MatLab
Changes in the two groups of participants in the study (19 PsA ; 12 HC ). * p-value < 0.005 ** p-value < 0.005. Quantification of small molecules was performed with a simplex approach based algorithm developed in-house and implemented in MatLab
 16s rRNA gene sequencing. Illumina MiSeq Regions V1-V2
16s rRNA gene sequencing. Illumina MiSeq Regions V1-V2
To cite this abstract in AMA style:
Miguens Blanco J, Selvarajah U, Lui Z, Mullish B, Alexander J, McDonald J, Abraham S, Marchesi J. Identification of New Associations Between Psoriatic Arthritis and the Gut Microbiota. the Mi-PART, a Phenomic Study [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/identification-of-new-associations-between-psoriatic-arthritis-and-the-gut-microbiota-the-mi-part-a-phenomic-study/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identification-of-new-associations-between-psoriatic-arthritis-and-the-gut-microbiota-the-mi-part-a-phenomic-study/
