Session Information
Date: Sunday, November 8, 2020
Title: Systemic Sclerosis & Related Disorders – Basic Science (1522–1526)
Session Type: Abstract Session
Session Time: 3:00PM-3:50PM
Background/Purpose: Genome-wide association study (GWAS) from the Genome Research in African American Scleroderma Patients (GRASP) cohort has identified the human leukocyte antigen (HLA) region as the strongest genetic risk factor in SSc. The functional roles of HLA variants in SSc susceptibility are largely unknown. We hypothesize that SSc-associated HLA variants overlap with one of the classes of cis-regulatory modules (CRMs) such as enhancer to regulate gene expression. CRMs can be mapped using DNase I hypersensitive site sequencing (DNase-Seq) or assay for transposase-accessible chromatin with high throughput sequencing (ATAC-Seq) giving information regarding chromatin accessibility. Thus, chromatin accessibility in the SSc-relevant cell types would be crucial in interpreting the function of these variants.
Methods: Genotyping data using the Illumina Multi-Ethnic Global array (MEGA) from the GRASP cohort were analyzed to identify the top HLA variant. Expression quantitative trait loci (eQTL) analysis was performed for the risk variants using RNA sequencing data from the Genotype-Tissue Expression (GTEx) project. Epstein-Barr virus-transformed B (EBV-B) cells with wild type, heterozygous and homozygous alleles for the most highly associated HLA variant were used to confirm the expression of HLA-II genes using RNA-sequencing. Protein expression was confirmed by flow cytometry and confocal microscopy. Chromatin accessibility at the variant position in EBV-B cells is being identified using ATAC-seq.
Results: The top variant in the African American GWAS was rs9469201, near the HLA-DQA1 gene with an odds ratio of 2.13 (P=4.5×10-20). eQTL analysis suggested decreased expression of HLA-DQA1/-DQB1 (in whole blood) and increased expression of HLA-DRB1 (in EBV-B cells) for the risk allele (Figure 1). This is particularly intriguing because we have previously reported two HLA-DRB1 alleles (HLA-DRB1*08:04 and -DRB1*11:02) as independent risk factors in African American SSc. The rs9469201 variant was found not to be in linkage disequilibrium with the HLA-DRB1*08:04 allele. Flow cytometry (Figure 2) and confocal microscopy (Figure 3) confirmed the expression of HLA-DRB1 and HLA-DQB1. ATAC-Seq analysis of the EBV-B cells is currently ongoing.
Conclusion: Our preliminary findings suggest that peptide presentation by HLA molecules as well as the relative expression of the specific HLA molecule presenting the peptide are both important in SSc pathogenesis. Future directions will involve CRISPR/Cas9 based deletion of CRMs-with overlapping HLA variants, and subsequent evaluation of the changes in HLA-II gene expression. Transcription factor binding at the HLA variant site will be accessed using chromatin immunoprecipitation with sequencing (ChIP-seq) in EBV-B cells. This study will elucidate the functional role of the HLA variants in allele-specific HLA-II gene expression in SSc.
 Figure 1. GTEx HLA-DRB1, HLA-DQA1 and HLA-DQB1 expression by rs9469201 variant
Figure 1. GTEx HLA-DRB1, HLA-DQA1 and HLA-DQB1 expression by rs9469201 variant
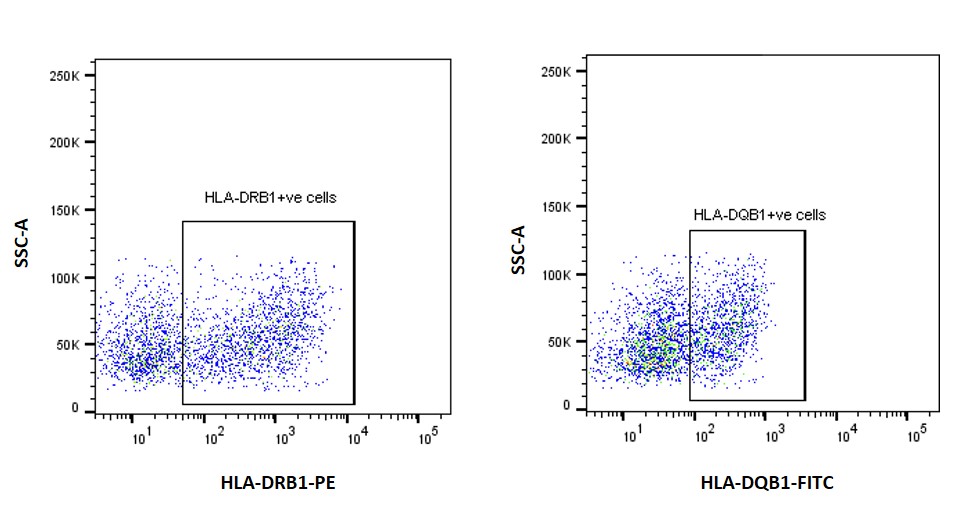 Figure 2. Flow cytometry plots for EBV-B cells with wild type allele stained for HLA-DRB1 and HLA-DQB1 protein using fluorochrome conjugated antibodies.
Figure 2. Flow cytometry plots for EBV-B cells with wild type allele stained for HLA-DRB1 and HLA-DQB1 protein using fluorochrome conjugated antibodies.
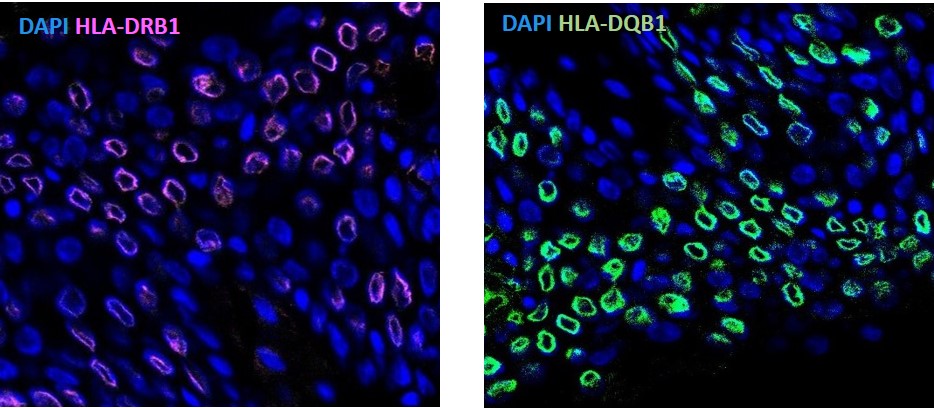 Figure 3. Confocal microscopy image for HLA-DRB1 and HLA-DQB1 stained cells for the wild type allele.
Figure 3. Confocal microscopy image for HLA-DRB1 and HLA-DQB1 stained cells for the wild type allele.
To cite this abstract in AMA style:
Kaundal U, Hartman J, Borden C, Wang J, Shah A, Mayes M, Doumatey A, Bentley A, Shriner D, Domsic R, Medsger T, Ramos P, Silver R, Steen V, Varga J, Hsu V, Saketkoo L, Schiopu E, Khanna D, Gordon J, Criswell L, Gladue H, Derk C, Bernstein E, Bridges S, Shanmugam V, Kolstad K, Chung L, Kafaja S, Jan R, Trojanowski M, Goldberg A, Korman B, Hinchcliff M, Chandrasekharappa S, Gadina M, Randazzo D, Dell'Orso S, Adeyemo A, Rotimi C, Remmers E, Wigley F, Casellas R, Kastner D, Boin F, Gourh P. Intergenic HLA Variants in African American Patients with Systemic Sclerosis Regulate Expression of HLA-DRB1 [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/intergenic-hla-variants-in-african-american-patients-with-systemic-sclerosis-regulate-expression-of-hla-drb1/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/intergenic-hla-variants-in-african-american-patients-with-systemic-sclerosis-regulate-expression-of-hla-drb1/
