Session Information
Date: Sunday, November 8, 2020
Title: Epidemiology & Public Health Poster III: Inflammatory Rheumatic Disease
Session Type: Poster Session C
Session Time: 9:00AM-11:00AM
Background/Purpose: Missing data in clinical epidemiological researches violate the intention to treat principle, reduce statistical power and can induce bias if they are related to patient’s response to treatment. In multiple imputation (MI), covariates are included in the imputation equation to predict the values of missing data. The purpose of this study is to find the best approach to estimate and impute the missing values in Kuwait Registry for Rheumatic Diseases (KRRD) patients data.
Methods: A number of methods were implemented for dealing with missing data. These included Multivariate imputation by chained equations (MICE), K-Nearest Neighbors (KNN), Bayesian Principal Component Analysis (BPCA), EM with Bootstrapping (Amelia II), Sequential Random Forest (MissForest) and mean imputation. Choosing the best imputation method was judged by the minimum scores of Root Mean Square Error (RMSE), Mean Absolute Error (MAE) and Kolmogorov–Smirnov D test statistic (KS) between the imputed datapoints and the original datapoints that were subsequently sat to missing.
Results: A total of 1,685 rheumatoid arthritis (RA) patients and 10,613 hospital visits were included in the registry. Among them, we found a number of variables that had missing values exceeding 5% of the total values. These included duration of RA (13.0%), smoking history (26.3%), rheumatoid factor (7.93%), anti-citrullinated peptide antibodies (20.5%), anti-nuclear antibodies (20.4%), sicca symptoms (19.2%), family history of a rheumatic disease (28.5%), steroid therapy (5.94%), ESR (5.16%), CRP (22.9%) and SDAI (38.0%), The results showed that among the methods used, MissForest gave the highest level of accuracy to estimate the missing values. It had the least imputation errors for both continuous and categorical variables at each frequency of missingness and it had the smallest prediction differences when the models used imputed laboratory values. In both data sets, MICE had the second least imputation errors and prediction differences, followed by KNN and mean imputation.
Conclusion: MissForest is a highly accurate method of imputation for missing data in KRRD and outperforms other common imputation techniques in terms of imputation error and maintenance of predictive ability with imputed values in clinical predictive models. This approach can be used in registries to improve the accuracy of data, including the ones for rheumatoid arthritis patients.
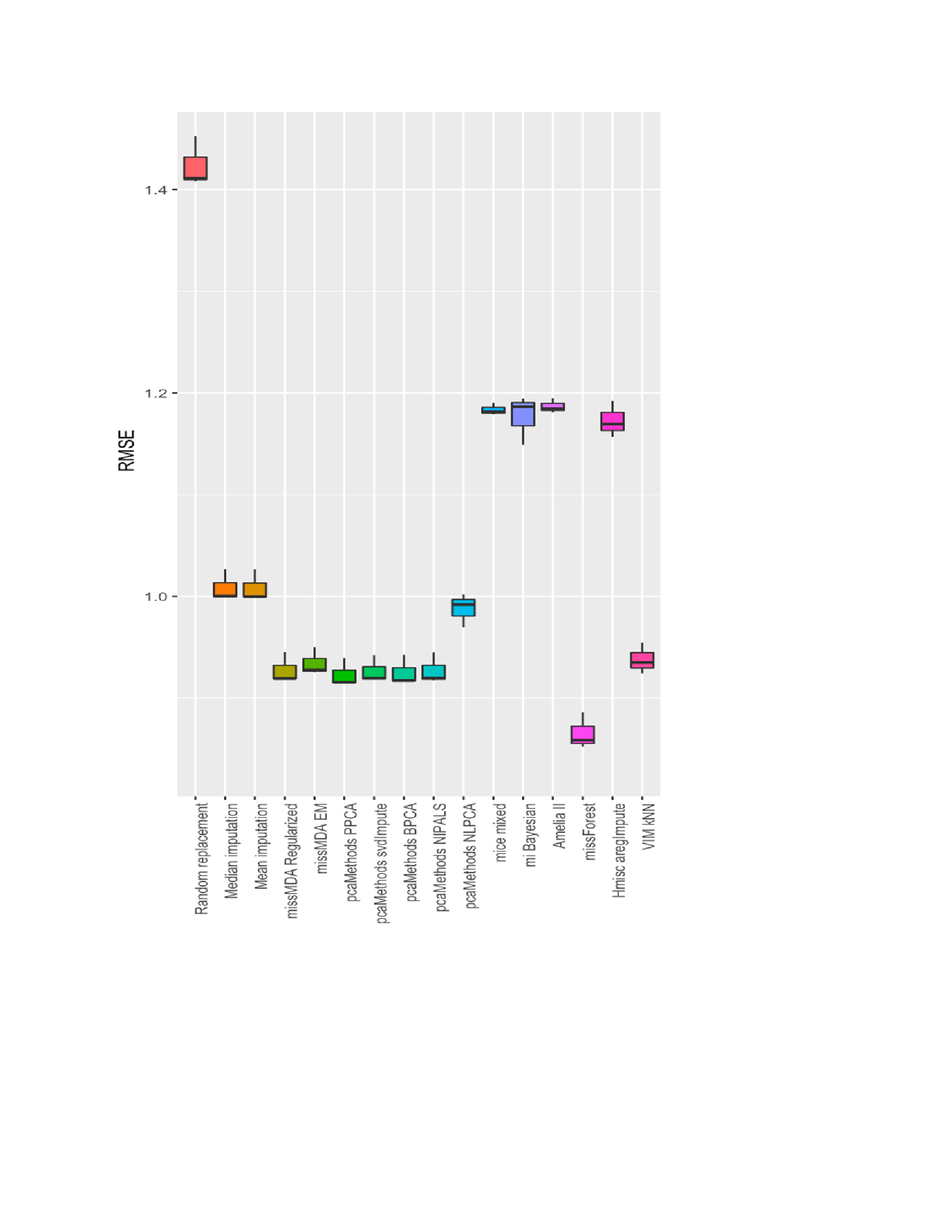 Root mean square error (RMSE) of various missing data imputation methods, showing that MissForest method has the lowest error to predict the missing information in KRRD data
Root mean square error (RMSE) of various missing data imputation methods, showing that MissForest method has the lowest error to predict the missing information in KRRD data
To cite this abstract in AMA style:
Al-Saber A, Al-Herz A, Pan J, Saleh K, Al-Awadhi A, Al-Kandari W, Hasan E, Ghanem A, Hussain M, Ali Y, Nahar E, Alenizi A, Hayat S, Abutiban F, Aldei A, Alkadi A, Alhajeri H, Behbehani H, Alhadhood N, Mokaddem K, Khadrawy A, Fazal A, Zaman A, Mazloum G, Bartella Y, Hamed S, Alsouk R. Missing Data and Multiple Imputation in Rheumatoid Arthritis Registries Using Sequential Random Forest Method [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/missing-data-and-multiple-imputation-in-rheumatoid-arthritis-registries-using-sequential-random-forest-method/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/missing-data-and-multiple-imputation-in-rheumatoid-arthritis-registries-using-sequential-random-forest-method/
