Session Information
Session Type: Abstract Session
Session Time: 3:00PM-3:50PM
Background/Purpose: Biomedical research uses many statistical/bioinformatics tools to find genes or proteins differentially expressed between patient group of interest. However, biological elements are related to one another in a complex network. Understanding those networks is vital to give results meaning in a disease context.
Treatment-naïve early rheumatoid arthritis (RA) patients have been described to have different histological pathotypes (lympho-myeloid, diffuse-myeloid, pauci-immune fibroid), with corresponding molecular signatures, associated with response to conventional synthetic Disease-Modifying Anti-Rheumatic Drugs (csDMARDs) [Lewis et al. 2019, Cell reports]. Purpose of this study was to use a novel pathway analysis approach to determine which gene networks are specifically activated in patients with different pathotypes or different response to csDMARDs.
Methods: We analysed RNA-Sequencing of 90 patients with early treatment-naïve RA fulfilling the 2010 ACR/EULAR Criteria recruited into the Pathobiology of Early Arthritis Cohort (PEAC). A detailed network of biological interactions was built starting from the KEGG pathway repository and enriched with miRNA interactions from miRTarBase and miRecords. Interactions between transcription factors (TFs) and miRNAs were added from TransmiR.
For each category, the most active links between genes, protein, metabolites and miRNA were selected based on their RNA-Sequencing expression values. Links redundantly active in two or more categories were removed. The resulting subnetworks show interactions that are active in the selected category only.
Results: The interactions networks found for the three pathotypes showed a gene level confirmation of the above taxonomy. The lympho-myeloid pathotype network (373 activator links, 96 inhibitor links) is characterized by lymphocyte activation with NFKB and mTOR signalling and recruitment via chemokines (for example CCL5, CCL2 and CCL19). We found IL3R signalling and cell survival networks to be central in the diffuse-myeloid pathotype (20 activator links, 5 inhibitor links), while in a pauci-immune fibroid environment (430 activator links, 99 inhibitor links) remodelling of the extracellular matrix and vasculature was prominent (Fig. 1).
On one hand, patients who had a good EULAR response to csDMARDs showed cell survival networks with MAP kinase and STAT signalling around BCL2 (Fig. 2). On the other hand, non-response was linked to T cell genes and lymphocyte attraction via chemokine signalling (Fig.3).
Conclusion: Our novel approach of investigating gene networks leads to a deeper understanding of differences in disease pathotypes and response to treatment. Allowing greater insight into underlying mechanisms, it will facilitate more targeted treatment.
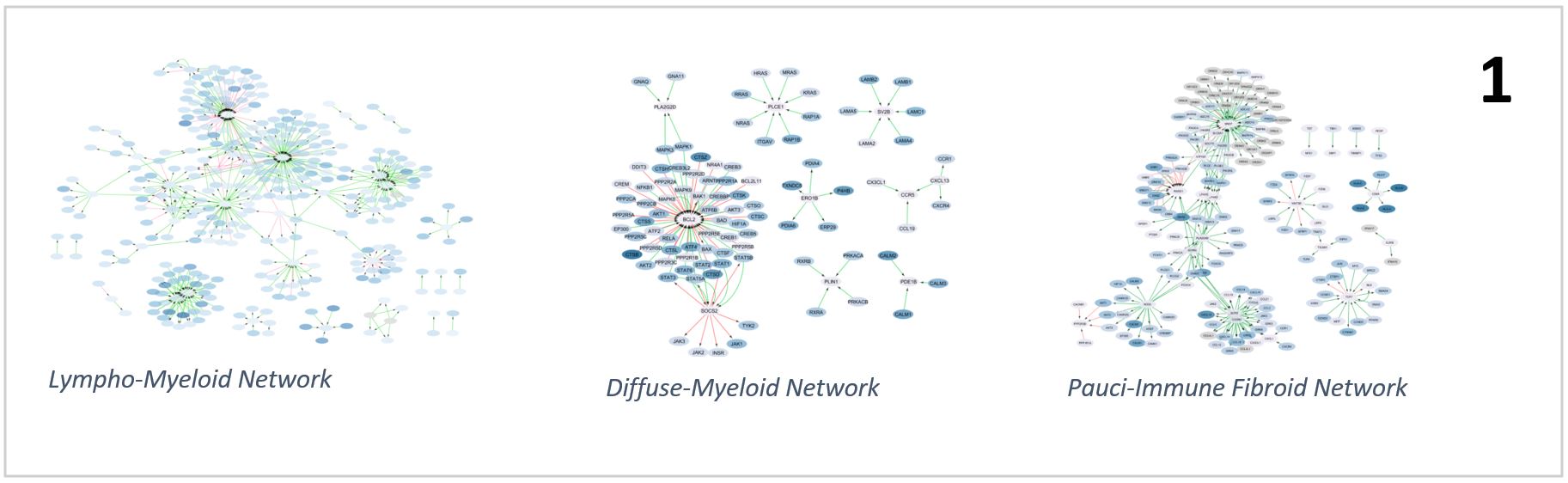 Pathotypes networks overview showing different number of nodes, edges and topology across the three groups.
Pathotypes networks overview showing different number of nodes, edges and topology across the three groups.
 Cell survival genes in patients responding to Disease-modifying anti-rheumatic drug (DMARD) treatment.
Cell survival genes in patients responding to Disease-modifying anti-rheumatic drug (DMARD) treatment.
 Chemokine and T cell cluster genes activated in patients not responding to DMARD treatment.
Chemokine and T cell cluster genes activated in patients not responding to DMARD treatment.
To cite this abstract in AMA style:
Sciacca E, Surace A, Lewis M, Alaimo S, Latora V, Rivellese F, Pulvirenti A, Ferro A, Pitzalis C. Novel Network Tool Highlights Key Features Associated with Disease Pathotypes and Response to Treatment in Early Rheumatoid Arthritis [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/novel-network-tool-highlights-key-features-associated-with-disease-pathotypes-and-response-to-treatment-in-early-rheumatoid-arthritis/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/novel-network-tool-highlights-key-features-associated-with-disease-pathotypes-and-response-to-treatment-in-early-rheumatoid-arthritis/
