Session Information
Session Type: Abstract Session
Session Time: 5:00PM-5:50PM
Background/Purpose: Previous studies demonstrated that tumor necrosis factor transgenic (TNF-Tg) mice with inflammatory arthritis have damaged lymphatic muscle cells (LMCs) and eventual loss of popliteal lymphatic vessel (PLV) contractions associated with severe joint disease.1 Anti-TNF therapy in flaring TNF-Tg mice recovers these PLV-LMCs and PLV contractions concomitant with amelioration of arthritis.1 As this regenerative process is critical for resumption of joint function, knowledge regarding PLV-LMC progenitor origins and physiology is needed. Surprisingly, LMCs express an enigmatic actin profile with co-existing striated and smooth muscle actin isoforms,2 which is incompatible with the differentiation of known muscle progenitors. To determine PLV-LMC origin, we tested the hypothesis that PLV-LMCs are derived from previously characterized striated or vascular smooth muscle cell (VSMC) progenitors via lineage tracing studies in mice. We also utilized these lineage tracing models to specifically isolate PLV-LMCs and VSMCs for comparative single cell RNA sequencing (scRNAseq) transcriptomic analysis.
1. Bouta et al. 2018. Nature Reviews Rheumatology. 14(2):94-106.
2. Muthuchamy et al. 2003. The FASEB Journal. 17(8):920-922.
Methods: Constitutive Cre and tamoxifen-inducible CreER models were crossed into Ai9tdTomato reporter lines for lineage tracing of PLV-LMCs during neonatal development. Induction of the CreER was achieved with 0.1 mg/g of tamoxifen administered intraperitoneal on postnatal days 10 – 13 (P10 – 13). All cohorts were sacrificed at both P21 and/or after 6-weeks. Pax7Cre and MyoDiCre animals were used to study striated muscle cell origin, while Prrx1Cre/CreER and NG2Cre/CreER mice were used to trace VSMC progenitors by tdTomato (tdT) expression visualized by ex vivo whole mount immunofluorescent microscopy of PLVs. The NG2Cre;tdT model was also utilized for fluorescence activated cell sorting (FACS) of tdT+ PLV-LMCs and VSMCs with downstream scRNAseq analysis on a 10X genomics platform.
Genes: Pax7 = Paired Box Protein 7; MyoD = Myogenic Differentiation Protein 1; Prrx1 = Paired Related Homeobox 1; Neural Glial Antigen 2 = NG2
Results: We demonstrate that PLV-LMCs originate from Pax7–/MyoD–/Prrx1+/NG2+ progenitors prior to P10, and from previously unknown Pax7–/MyoD–/Prrx1+/NG2– progenitors during development after P10 (Figure 1). In addition, our preliminary scRNAseq results suggest that mature PLV-LMCs are transcriptionally distinct muscle cells (Figure 2).
Conclusion: This is the first study to describe the unique origin of PLV-LMC progenitors and to assess the distinct characterization of mature PLV-LMCs using a scRNAseq approach. Our analysis suggests that PLV-LMCs do not derive from striated muscle progenitors with Pax7- and MyoD-negativity. In addition, PLV-LMCs appear to originate from similar Prrx1+/NG2+ progenitors to VSMCs, but derive from distinct Prrx1+/NG2– progenitors prior to P10 to become transcriptionally unique muscle cells. Thus, future work elucidating the PLV-LMC lineage, and defining specific markers of PLV-LMCs, will catalyze the development of LMC targeted therapies for diseases with lymphatic involvement, such as inflammatory arthritis.
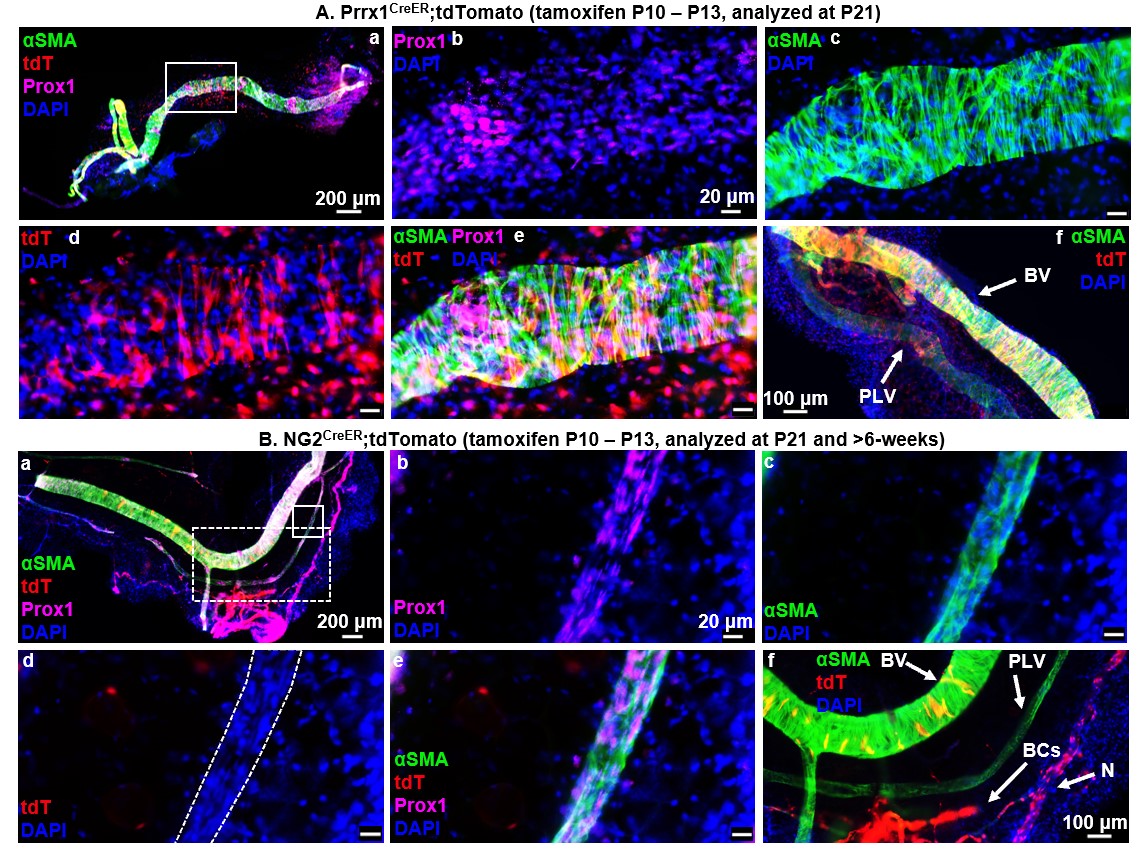 Figure 1. LMCs derive from a unique Prrx1+ and NG2- progenitor cell incorporated onto PLVs between P10 and P21. Multicolor fluorescent microscopy was performed on whole mount immunostained PLVs and adjacent blood vessels (BVs) from tamoxifen-inducible Prrx1CreER (n = 6, P21 depicted), and NG2CreER (n = 5, P90 depicted) reporter mice as described in Fig. 1. Representative low-magnification overlay images of all channels show the PLVs with a highlighted region of interest (white boxes) (A.a, B.a). High-magnification images of the region of interest show Prox1 immunostain of LECs (A.b, B.b), αSMA immunostain of LMCs (A.c, B.c), Cre-driven tdT expression (A.d, B.d), overlay of all channels (A.e, B.e), and positive control BV VSMCs (A.f, B.f), blood capillary (BC) pericytes, and peripheral nerves (N) all directly adjacent to PLVs (B.f). Lineage tracing of Prrx1CreER (A.a-f) mice demonstrates tdT expression in PLVs (A.d) that colocalizes with αSMA+ LMCs (A.e) along with successful Cre-recombination with tdT expression in positive control VSMCs, as expected (A.f). NG2CreER (B.a-f) shows absent tdT expression in PLVs (no red staining in outlined PLV) (B.d) with successful tamoxifen induced NG2-driven Cre-recombination depicted by tdT expression in BV VSMCs, BC pericytes, and peripheral nerves (N), as expected (B.f). Note the few yellow VSMCs on the BV, which indicates that these cells were added to the fully formed BV after tamoxifen treatment.
Figure 1. LMCs derive from a unique Prrx1+ and NG2- progenitor cell incorporated onto PLVs between P10 and P21. Multicolor fluorescent microscopy was performed on whole mount immunostained PLVs and adjacent blood vessels (BVs) from tamoxifen-inducible Prrx1CreER (n = 6, P21 depicted), and NG2CreER (n = 5, P90 depicted) reporter mice as described in Fig. 1. Representative low-magnification overlay images of all channels show the PLVs with a highlighted region of interest (white boxes) (A.a, B.a). High-magnification images of the region of interest show Prox1 immunostain of LECs (A.b, B.b), αSMA immunostain of LMCs (A.c, B.c), Cre-driven tdT expression (A.d, B.d), overlay of all channels (A.e, B.e), and positive control BV VSMCs (A.f, B.f), blood capillary (BC) pericytes, and peripheral nerves (N) all directly adjacent to PLVs (B.f). Lineage tracing of Prrx1CreER (A.a-f) mice demonstrates tdT expression in PLVs (A.d) that colocalizes with αSMA+ LMCs (A.e) along with successful Cre-recombination with tdT expression in positive control VSMCs, as expected (A.f). NG2CreER (B.a-f) shows absent tdT expression in PLVs (no red staining in outlined PLV) (B.d) with successful tamoxifen induced NG2-driven Cre-recombination depicted by tdT expression in BV VSMCs, BC pericytes, and peripheral nerves (N), as expected (B.f). Note the few yellow VSMCs on the BV, which indicates that these cells were added to the fully formed BV after tamoxifen treatment.
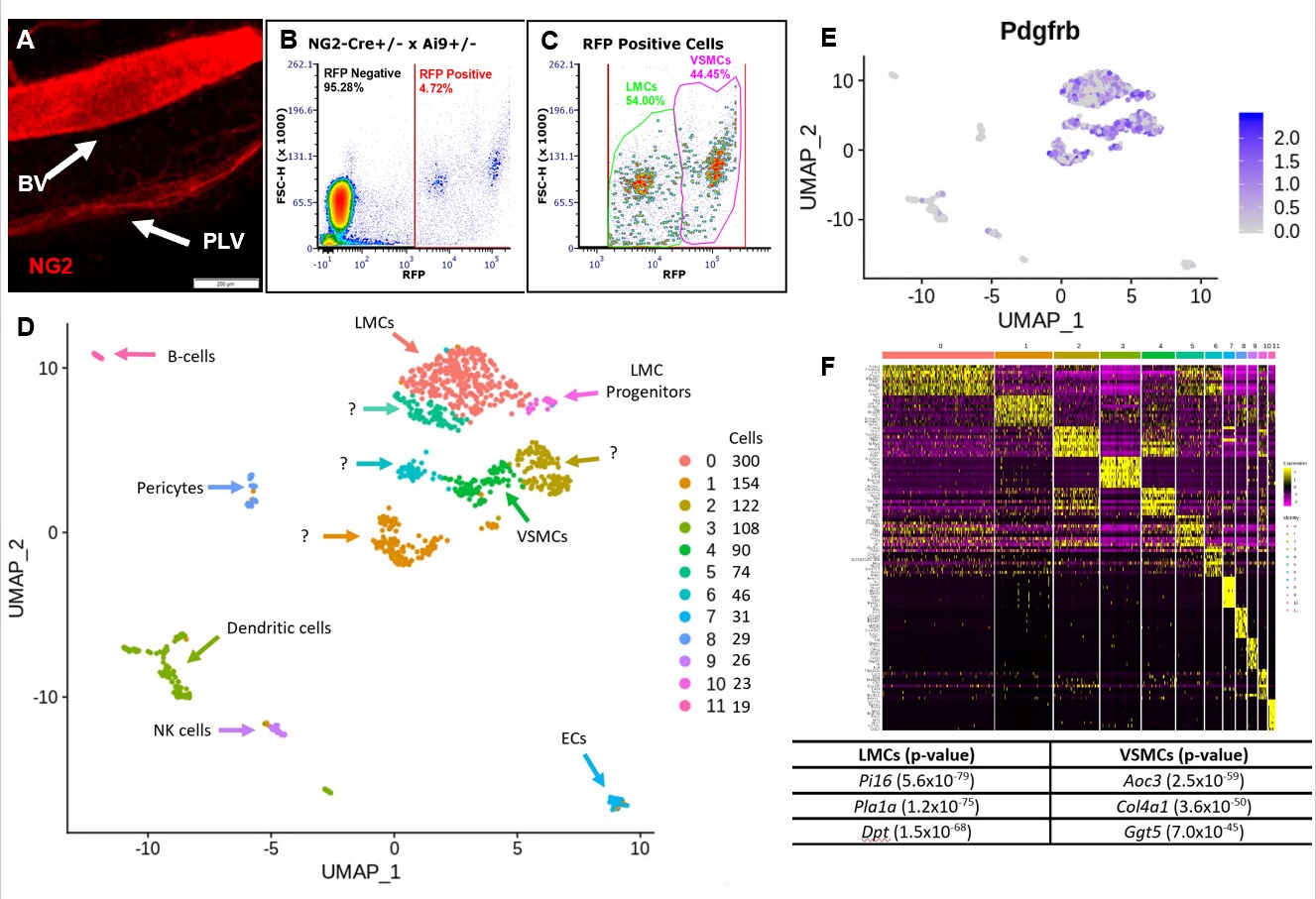 Figure 2. FACS and scRNAseq of tdT+ PLVs and BVs identifies LMC and VSMC populations. Whole mount fluorescent microscopy of PLVs and adjacent BV from NG2-Cre+/- x Ai9+/- mice revealed that both LMCs and VSMCs are RFP+, but VSMCs are much brighter than LMCs (MFI = 63.83 +/- 5.43 vs. 22.28 +/- 5.45 in arbitrary units; p < 0.05 n=5 vessels) (A). A single cell suspension was prepared by pooling the lateral saphenous vein and both adjacent PLVs from both hindlegs of 3 NG2-Cre+/- x Ai9+/- mice. The cells were analyzed by FACS with ~6,000 RFP+ events/animal (B), and revealed two distinct populations based on RFP signal intensity (C), presumed to be LMCs and VSMCs based on the whole mount studies. The tdT+ sample contained 1,504 viable cells and was analyzed using scRNAseq (93,099 reads of 2,142 genes/cell with 97.2% barcode validity). UMAP was used to embed the data into 2D space and an sNN clustering algorithm resolved 12 clusters (Clusters 0-11 with cells/cluster, right; Seurat, R). Clusters were annotated using Mouse Gene Atlas, KEGG, Wiki Pathways & GO Biological Process (D). High levels of Pdgfrb expression are seen in the clusters annotated as LMCs and VSMCs validating their identification as mesenchymal muscle cells (E). A heatmap showing the ten most differentially regulated genes between each cluster identified unique muscle-defining genes between LMCs and VSMCs (F).
Figure 2. FACS and scRNAseq of tdT+ PLVs and BVs identifies LMC and VSMC populations. Whole mount fluorescent microscopy of PLVs and adjacent BV from NG2-Cre+/- x Ai9+/- mice revealed that both LMCs and VSMCs are RFP+, but VSMCs are much brighter than LMCs (MFI = 63.83 +/- 5.43 vs. 22.28 +/- 5.45 in arbitrary units; p < 0.05 n=5 vessels) (A). A single cell suspension was prepared by pooling the lateral saphenous vein and both adjacent PLVs from both hindlegs of 3 NG2-Cre+/- x Ai9+/- mice. The cells were analyzed by FACS with ~6,000 RFP+ events/animal (B), and revealed two distinct populations based on RFP signal intensity (C), presumed to be LMCs and VSMCs based on the whole mount studies. The tdT+ sample contained 1,504 viable cells and was analyzed using scRNAseq (93,099 reads of 2,142 genes/cell with 97.2% barcode validity). UMAP was used to embed the data into 2D space and an sNN clustering algorithm resolved 12 clusters (Clusters 0-11 with cells/cluster, right; Seurat, R). Clusters were annotated using Mouse Gene Atlas, KEGG, Wiki Pathways & GO Biological Process (D). High levels of Pdgfrb expression are seen in the clusters annotated as LMCs and VSMCs validating their identification as mesenchymal muscle cells (E). A heatmap showing the ten most differentially regulated genes between each cluster identified unique muscle-defining genes between LMCs and VSMCs (F).
To cite this abstract in AMA style:
Kenney H, Bell R, Masters E, Xing L, Ritchlin C, Schwarz E. Investigating Murine Joint-Draining Lymphatics: Lineage Tracing and Single Cell RNA Sequencing Reveal Evidence That Popliteal Lymphatic Muscle Cells and Their Progenitors Represent Distinct Cell Types Divergent from Striated and Vascular Muscle Cells [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/investigating-murine-joint-draining-lymphatics-lineage-tracing-and-single-cell-rna-sequencing-reveal-evidence-that-popliteal-lymphatic-muscle-cells-and-their-progenitors-represent-distinct-cell-types/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/investigating-murine-joint-draining-lymphatics-lineage-tracing-and-single-cell-rna-sequencing-reveal-evidence-that-popliteal-lymphatic-muscle-cells-and-their-progenitors-represent-distinct-cell-types/
