Session Information
Session Type: Poster Session B
Session Time: 9:00AM-11:00AM
Background/Purpose: Epigenetic changes contribute to the pathogenesis of rheumatoid arthritis (RA) and a comprehensive epigenomic characterization of RA fibroblast-like synoviocytes (FLS) has recently been described. Nutrient transporters, with the SLC transporter families that contain 400 genes and 52 subfamilies among others, serve as ‘metabolic gate’ of cells by mediating the transport of a wide range of essential nutrients and metabolites such as glucose, amino acids, vitamins, neurotransmitters, and inorganic/metal ions. As previous studies indicate that metabolism is altered in RA FLS, we hypothesize that ChIP mark changes in nutrient transporters genes would correlate with differences in expression of these genes and would help to identify activated metabolic pathways.
Methods: ChIP-sequencing data, for six different ChIP marks (H3K4me1, H3K4me3, H3K9me3, H3K27ac, H3K27me3, H3K36me3), and DNA methylation from a publicly available data set from FLS derived from 11 RA patients and 11 OA patients were compared to identify regions with a difference in these histone modifications. Single nearest genes to regions of interest were then utilized for pathway analyses (REACTOME classification) to determine if particular cellular processes and pathways are associated with these chromatin changes. Pathways associated with nutrient transporters were enriched near ChIP mark changes commonly associated with active transcription (H3K4me1, H3K4me3, H3K27ac) with the use of the whole genome as a reference. To further elucidate these findings, single nearest genes associated with any ChIP mark change and transporters were utilized for a secondary pathway analysis. Changes in transcription of genes associated with the ChIP mark changes were assessed using RNA-sequencing data from the same cells. ATAC-Seq to study the open chromatin regions in FLS was also assessed.
Results: As per the unbiased pathway analysis, 21 pathways significantly associated with at least 3 either epigenetic marks, RNA-Seq or ATAC-Seq changes were associated with energy metabolism. “SLC-mediated transmembrane transport” was one of these pathways and associated with 4 changes. 34 different genes involved in nutrient transportation were associated with a change in at least 4 epigenetic marks (Figure 1). These genes were then used in a Reactome pathway analysis. Amino acids and oligopeptide SLC transporters (including SLC38A4, SLC38A1, SLC6A6, SLC7A8, SLC7A5, SLC7A11, SLC7A1, SLC1A4, SLC15A1, SLC15A4) was one of the top ranked pathways (adjusted p= 5.13e-13). Of note, most of these transporters are known to be glutamine transporters. Transporters for organic and metal ions were also heavily epigenetic regulated. Interestingly, ATAC-Seq analysis showed a significant enrichment of open chromatin regions near those genes.
Conclusion: This study of RA FLS demonstrates changes in epigenetic marks of genes related to nutrient transporters and suggests that metabolism is critical in RA pathogenesis and may be involved in the imprinted aggressive phenotype displayed by RA compared to OA FLS. Additionally, this dataset has the potential to identify RA-specific targets that can be used to develop novel therapeutic agents.
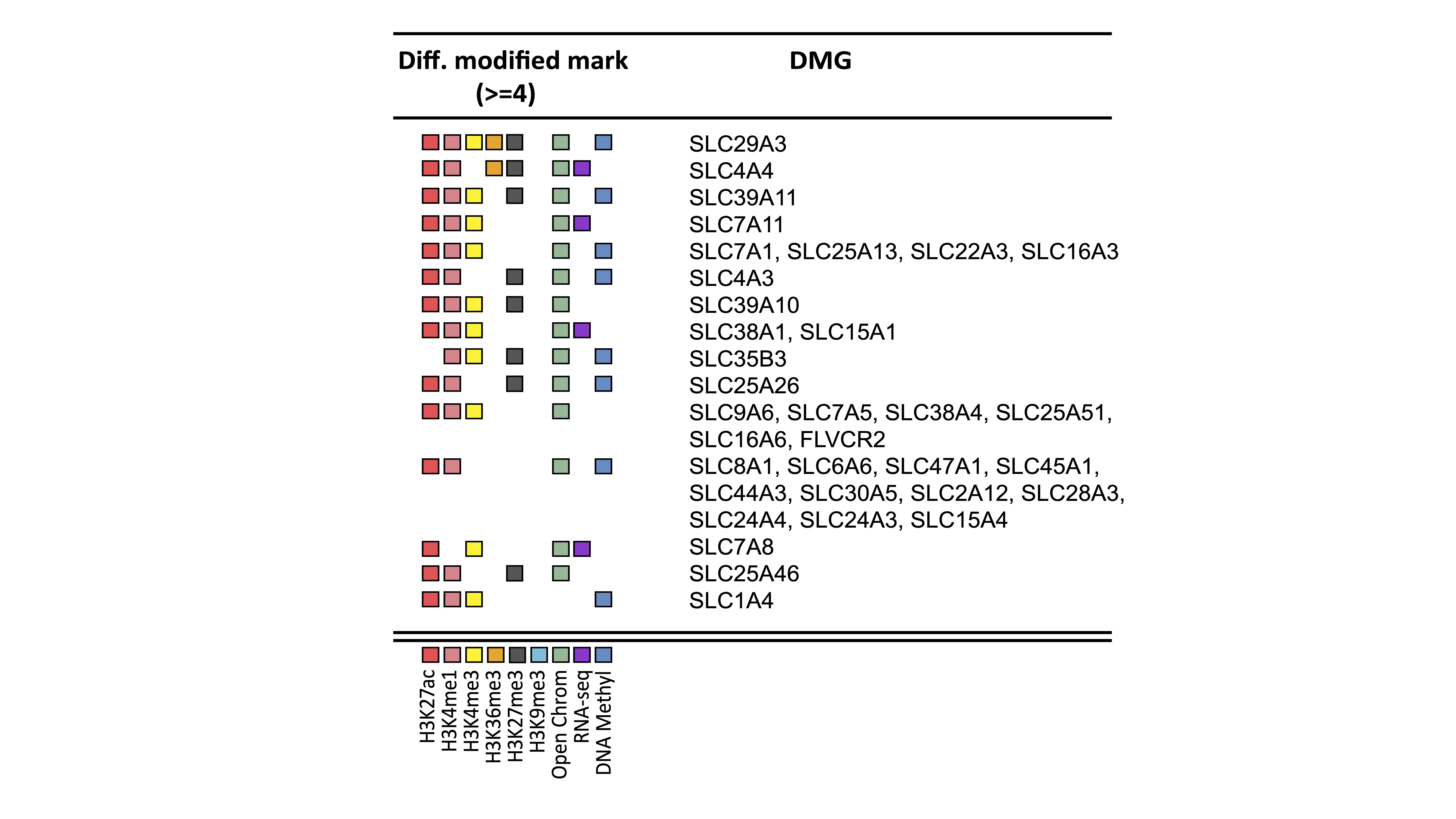 Figure 1: Shown the 34 differentially modified nutrient transport genes with differentiated modified marks.
Figure 1: Shown the 34 differentially modified nutrient transport genes with differentiated modified marks.
To cite this abstract in AMA style:
Pedersen B, Ai R, Torres A, Wang W, Firestein G, Guma M. Epigenetic Regulation of Metabolic Transporters in Rheumatoid Arthritis Fibroblast-Like Synoviocytes [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/epigenetic-regulation-of-metabolic-transporters-in-rheumatoid-arthritis-fibroblast-like-synoviocytes/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/epigenetic-regulation-of-metabolic-transporters-in-rheumatoid-arthritis-fibroblast-like-synoviocytes/
