Session Information
Date: Saturday, November 7, 2020
Title: RA – Diagnosis, Manifestations, & Outcomes Poster II: Biomarkers
Session Type: Poster Session B
Session Time: 9:00AM-11:00AM
Background/Purpose: Though targeted therapeutics have improved outcomes in Rheumatoid Arthritis (RA) treatment, the understanding of the underlying biological mechanisms that mediate inflammatory arthritis in RA is incomplete. We aimed to use unsupervised proteomics to identify serum proteins and active mechanistic networks that are associated with disease activity in RA.
Methods: Using the SOMAscan (slow off-rate modified aptamer) array, we generated quantitative levels of 1307 proteins in serum samples from 68 RA patients. Differentially expressed proteins between patients with high disease activity (HDA, DAS28 > 3.2) relative to low disease activity (LDA) were determined using linear modeling and visualized with hierarchical clustering. Spearman correlation was used to determine the relationship between individual proteins and DAS28, a disease activity score widely use in RA. Correlation adjusted mean rank (CAMERA) competitive gene set testing was performed to identify pathway enrichment while accounting for inter-gene correlation. Network analysis was then performed using Clusterprofiler and gene-concept network.
Results: 50% (n = 34) of the cohort were classified as HDA DAS28 (median 4.49 +/- 0.89) while 50% (n = 34) were LDA DAS28 (median 2.68, +/- 0.82). Principle components analysis based on expression of all proteins did not clearly separate HDA from LDA patients. CST7 (Fold Change (FC) = 6.2)) , ELANE (FC = 1.1), S100A12 (FC = 1.9, p < 0.001) were all highly upregulated in HDA, while CTLA4 (FC = 11.1) was the highest upregulated protein in LDA. Correlation between DAS28 and SERPINA3 (R = 0.53, p = 2.4 x 10-6) and S100A12 (R = 0.46, p = 8.6 x 10-5) were numerically higher than CRP (0.42, p = 0.0003). Gene-concept plotting linked extracellular matrix disassembly, endopeptidase activity, platelet degranulation, and leukocyte migration/antimicrobial humoral response with 19 upregulated proteins. Gene set enrichment identified Immature Neutrophil upregulation as one of the highest enriched gene networks in HDA (p = 1.49 X 10-4).
Conclusion: Our rich proteomic dataset provides a foundation for understanding of the underlying biological processes that underpin active RA. In particular, we identified proteins that implicate systemic neutrophil degranulation as a highly active process in RA, which may link systemic inflammation and tissue destruction in these individuals.
 Figure 1: Volcano plot depicting fold differences between low disease activity RA patients (LDA) and high disease activity RA patients (HDA), coloured by magnitude of fold change.
Figure 1: Volcano plot depicting fold differences between low disease activity RA patients (LDA) and high disease activity RA patients (HDA), coloured by magnitude of fold change.
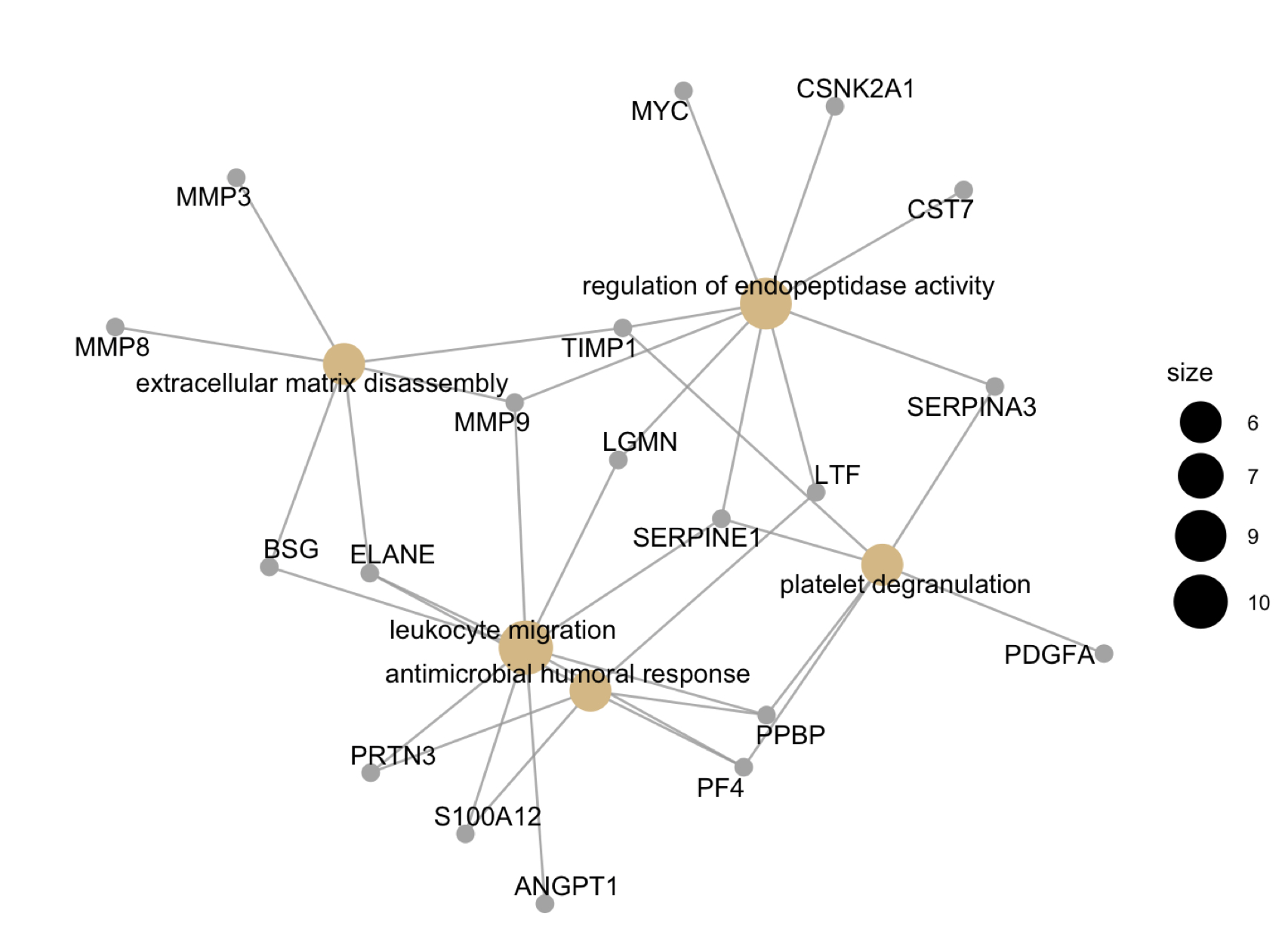 Figure 2: Gene network of protein interactions linked with GO biological processes using gene-concept plotting. Size of nodes are reflective of member inclusion. All protein members are upregulated in high disease activity relative to low disease activity.
Figure 2: Gene network of protein interactions linked with GO biological processes using gene-concept plotting. Size of nodes are reflective of member inclusion. All protein members are upregulated in high disease activity relative to low disease activity.
To cite this abstract in AMA style:
O'Neil L, Anaparti V, Wiens D, Smolik I, Meng X, El-Gabalawy H. Serum Proteomics Implicates Neutrophil Degranulation in Rheumatoid Arthritis Disease Activity [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/serum-proteomics-implicates-neutrophil-degranulation-in-rheumatoid-arthritis-disease-activity/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/serum-proteomics-implicates-neutrophil-degranulation-in-rheumatoid-arthritis-disease-activity/
