Session Information
Session Type: Poster Session A
Session Time: 9:00AM-11:00AM
Background/Purpose: Juvenile localized scleroderma (jLS) is an autoimmune disease of the skin and underlying tissue characterized by an early inflammatory infiltrate followed by fibrosis and collagen deposition leading to atrophy. Uncontrolled jLS results in significant disfigurement and functional disability. Our goal was to determine the transcriptome within inflammatory and fibrotic LS tissue vs. healthy controls and identify potential molecular targets using RNA sequencing (RNAseq). Differentially expressed genes (DEGs) identified in jLS patients were compared to histopathological features to determine correlation and were used to cluster LS patients according to common expression pathways.
Methods: RNAseq was performed on paraffin-embedded skin (n=28 jLS, n=10 pediatric healthy) using the Illumina HTS and TrueSeq Access library preparation. Paired-end RNAseq data was aligned using STAR, corrected for batch effect and analyzed for DEGs using DESeq2. Genes were analyzed using DEG cutoffs of log fold change > ±2.0, adjusted p< 0.05, and a false discovery rate (FDR) cutoff of < 0.05 for Reactome and enrichment software (GSEA©). Standardized scoring was developed for inflammation and collagen deposition and was completed between 2 blinded pathologists. Spearman’s correlation was used to determine significance between DEGs and histology scoring.
Results: We found 589 significant DEGs between jLS vs. pediatric healthy controls with the above cutoffs. Hierarchical clustering using complete linkage of Euclidean distance demonstrated three distinct jLS groupings; Group 1, Group 2 and Healthy-Like (Figure 1). In Group 1, 61 DEGs were identified which associate with interferon gamma signaling, MHC class II antigen presentation, and TCR signaling (p< 0.01). Specifically, five are HLA class II genes, HLA-DQA1, HLA-DQB1, HLA-DRB5, HLA-DPA1, and HLA-DRB1 (p< 0.01, FC >2, FDR< 0.05). Degree of inflammatory cell infiltrates significantly correlated with HLA-DPB1, HLA-DQA2, HLA-DRA, and STAT1 (rs > 0.5, p< 0.01). Collagen thickness in upper reticular dermis had a positive correlation with HLA-DQA1 (rs=0.46, p=0.03). In Group 2, 76 DEGs were identified, which associate with pathways of FGFR1 amplification, collagen formation and keratinization (p< 0.05).
Conclusion: The identified groupings of jLS patients showed two distinct genetic signatures, one with upregulated inflammatory-related pathways, which corresponded to inflammatory infiltrate score, and a second group with upregulated fibrosis-related pathways. HLA Class II gene upregulation was observed within the inflammatory group, which has also been described for systemic sclerosis. Interestingly also the immune-phenotype groups did not directly correspond with the patients clinical phenotype. Further investigation into the relationship and functions of these genes in jLS tissue is underway.
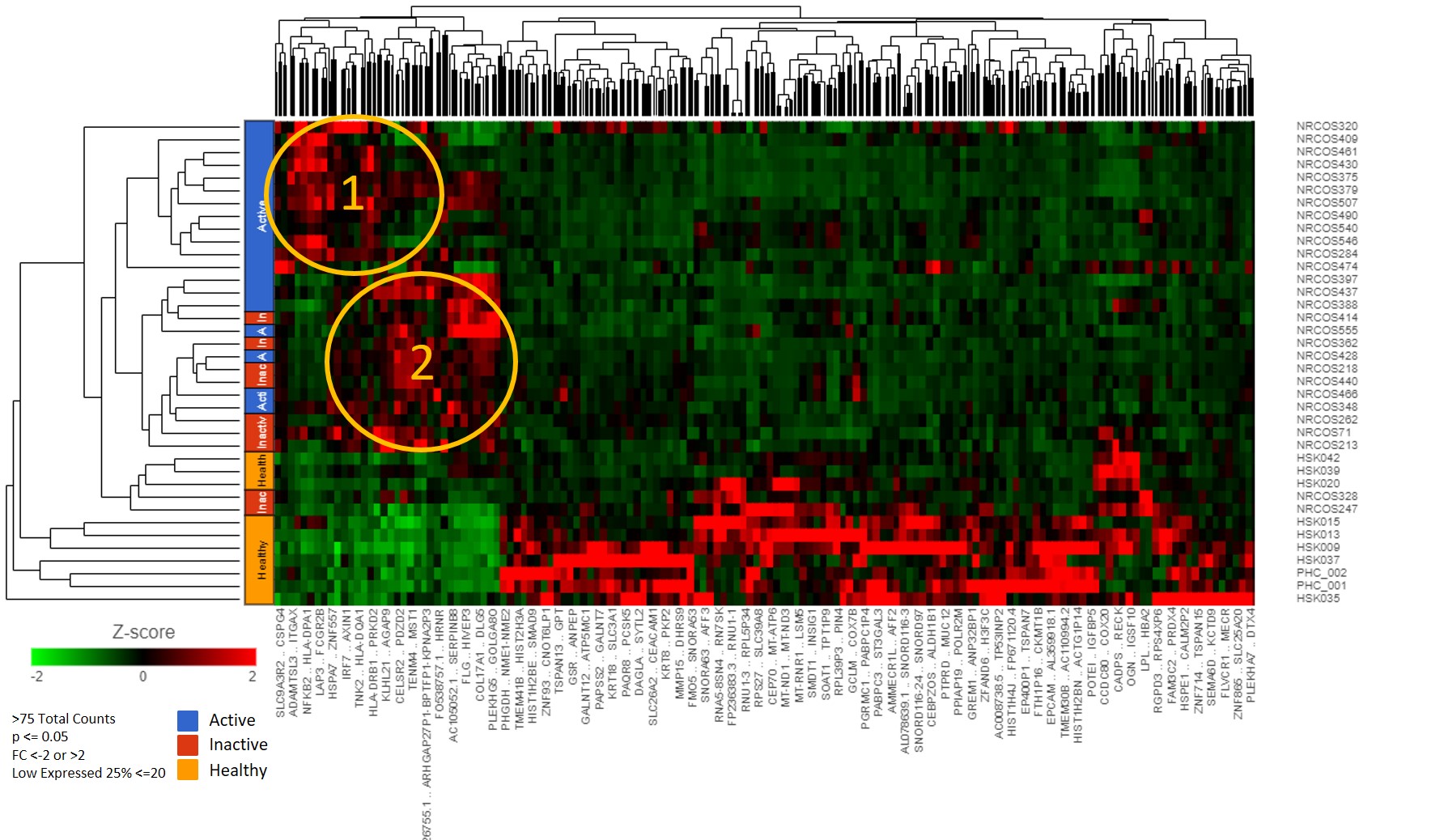 Figure 1: Hierarchical Clustering using complete linkage of Euclidean Distance
Figure 1: Hierarchical Clustering using complete linkage of Euclidean Distance
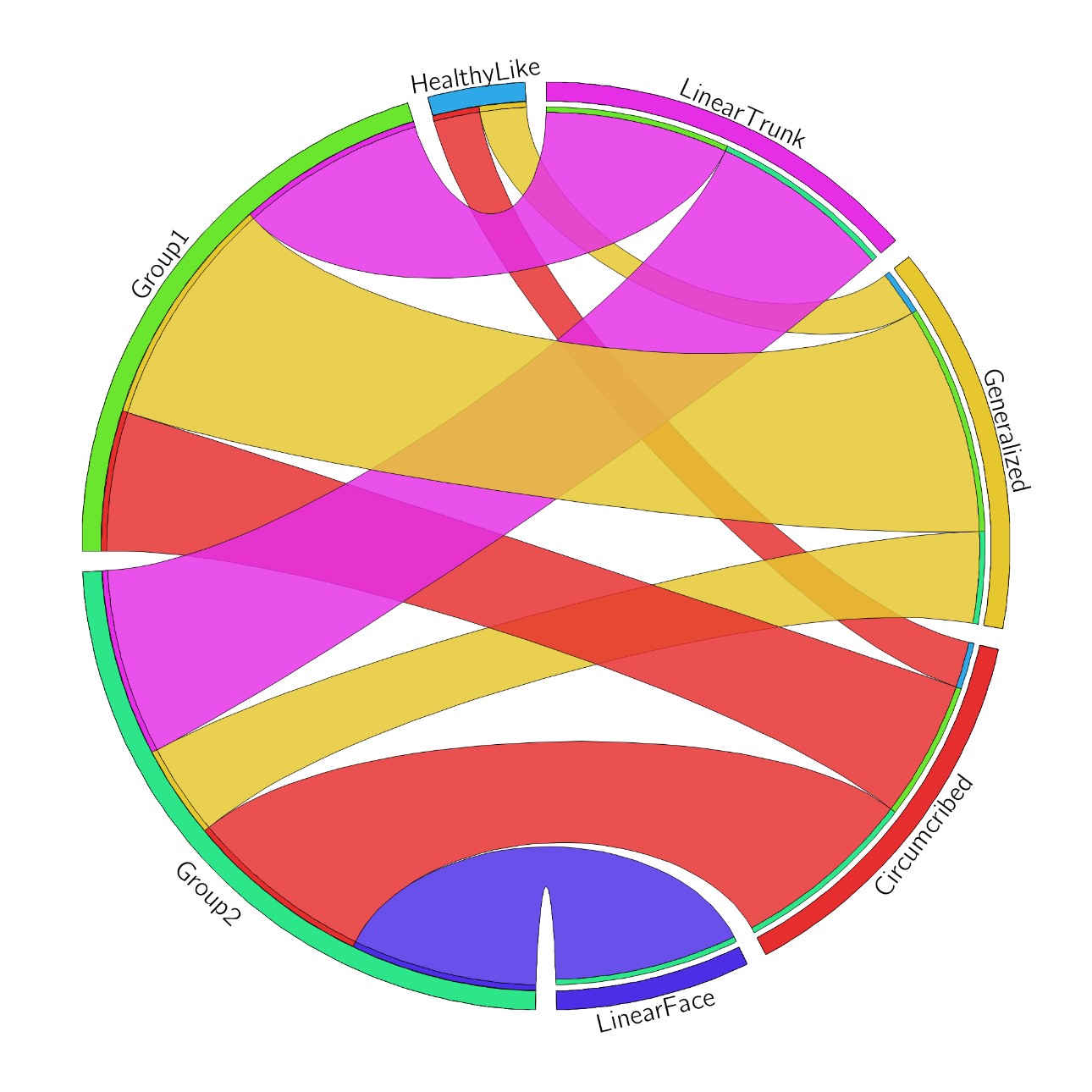 Figure 2: Circos plot comparing clinical LS subtype to immune-phenotype clustering.
Figure 2: Circos plot comparing clinical LS subtype to immune-phenotype clustering.
To cite this abstract in AMA style:
Schutt C, Mirizio E, Schollaert-Fitch K, Salgado C, Reyes-Mugica M, Torok K. Identifying Immuno-phenotypes in Juvenile Localized Scleroderma with RNA Sequencing [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/identifying-immuno-phenotypes-in-juvenile-localized-scleroderma-with-rna-sequencing/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identifying-immuno-phenotypes-in-juvenile-localized-scleroderma-with-rna-sequencing/
