Session Information
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Juvenile systemic sclerosis (jSSc) is a rare chronic autoimmune disorder characterized by skin thickening and multisystem organ involvement, leading to significant morbidity. The pathogenesis of jSSc is understudied. Whole blood transcriptome profiling can provide insights into genetic activity and identify key biomarkers involved in the disease. This study aims to reveal similarities between blood and skin transcriptomes, contributing to the understanding of jSSc pathology in children. To evaluate the peripheral blood transcriptome signature of jSSc patients compared to healthy pediatric controls (HC) using bulk RNA sequencing (RNA-Seq) and differential expression gene (DEG) analysis.
Methods: RNA-Seq was performed on RNA extracted from peripheral blood samples of jSSc patients (n=22) and age/sex-matched healthy controls (n=17). RNA was prepared for sequencing using Illumina Stranded Total Library Prep with Ribo-Zero Plus and sequenced on an Illumina NovaSeq 6000. Data analysis was conducted using Partek software and DESeq2. DEGs were identified based on a log2 fold change with a cutoff (FDR < 0.05) and fold change of ≤1.5 or ≥ 1.5. PCA and t-SNE were used to visualize gene expression clustering. Further analysis identified pathway enrichment and dysregulated pathways.
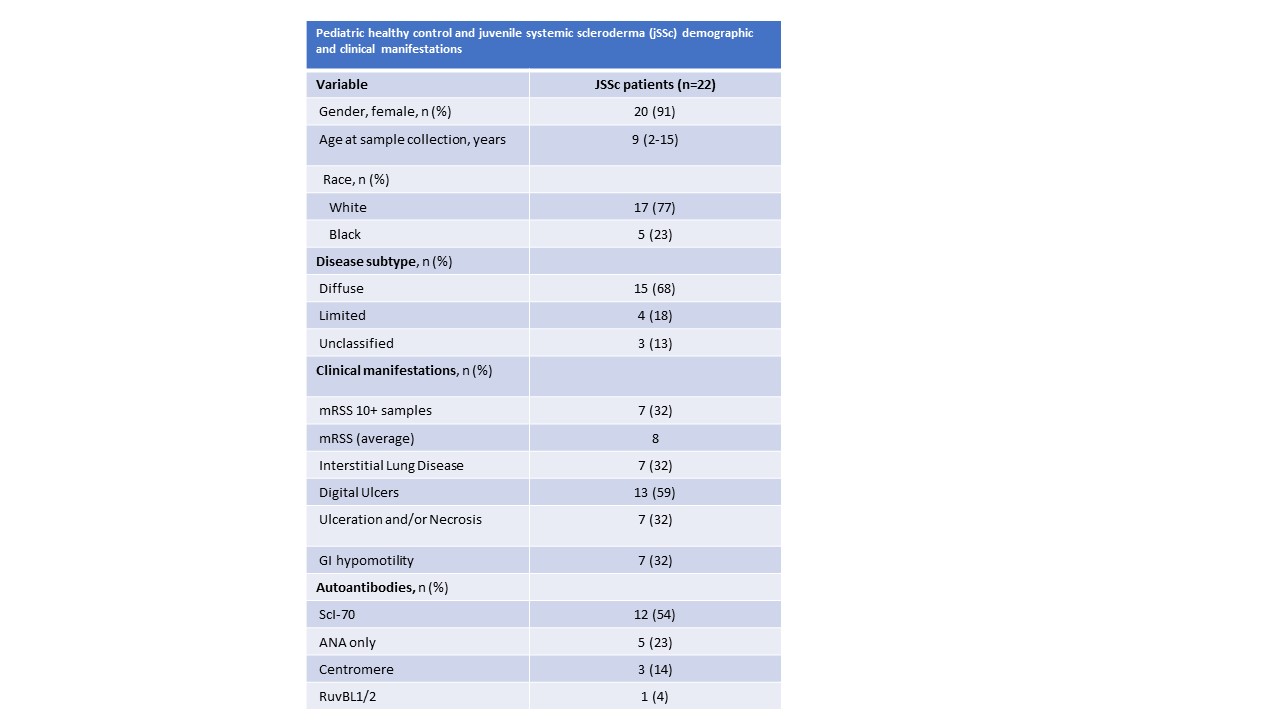
Results: The average age of onset for jSSc subjects was 9 years old (range 2-15), and healthy controls was 13 years old (range 5-20), both with female predominance, 90% and 75% respectively. The jSSc subjects were mostly diffuse cutaneous subtype with a mRSS of 8, two-thirds had digital ulcers and one-third had interstitial lung disease (Table 1). The jSSc patients’ RNA expression separate nicely from the HC (data not shown).
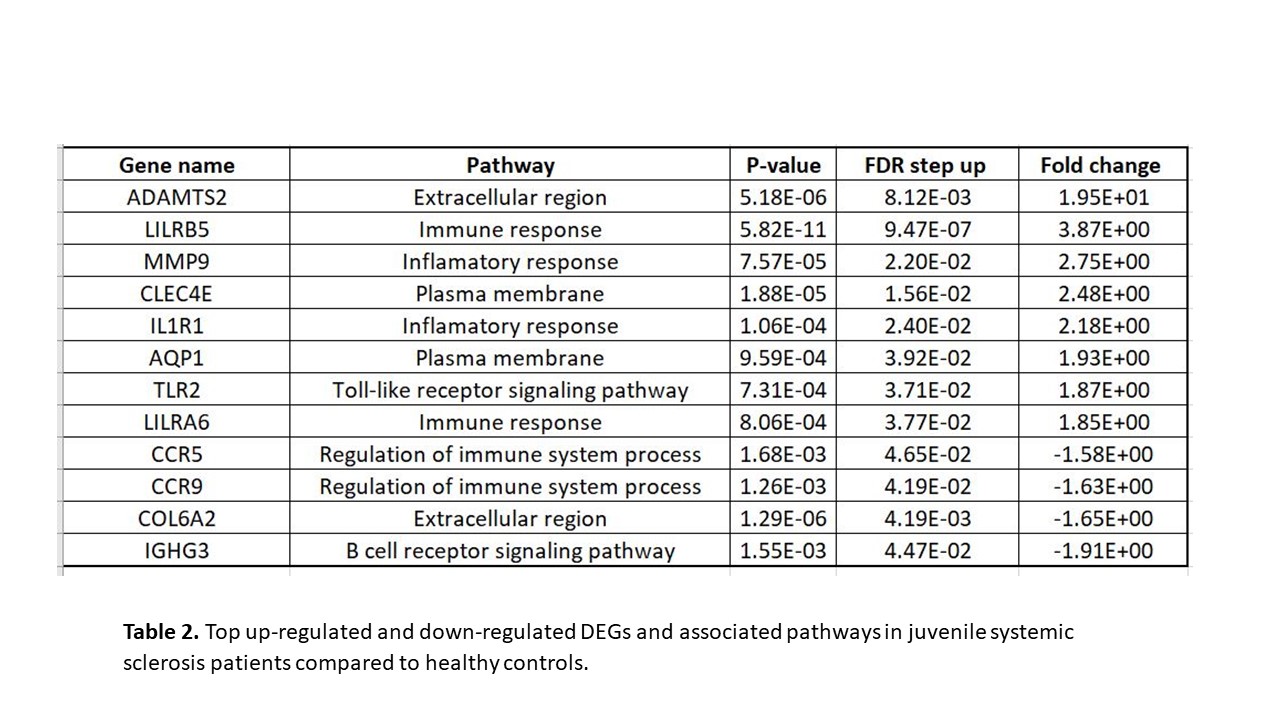
A total of 263 DEGs were identified when comparing jSSc with HC, with 200 upregulated. Key upregulated genes included ADAMTS2, AQP1, TREM1, and CLEC4E, which are involved in collagen and extracellular matrix (ECM) formation and play critical roles in connective tissue disorders (Table 2). Upregulated interleukin receptor (IL1R1) and toll-like receptors (TLRs) suggest their potential as blood biomarkers for disease severity. The downregulated COL6A2 gene, which encodes for Type VI collagen, indicates an imbalance in ECM production and degradation, contributing to fibrosis in jSSc. Gene set enrichment analysis (GSEA) revealed dysregulated pathways related to immune and inflammatory responses (e.g., MHC class II biosynthetic process, cytokine production regulation, innate and adaptive immune signaling) and collagen regulation (e.g., plasma membrane, collagen metabolic process).
Conclusion: The peripheral blood transcriptome of jSSc patients shows differentially expressed genes involved in ECM formation and activated pathways associated with inflammatory and immune responses, underscoring their roles in the pathogenesis of jSSc. These findings provide valuable insights into the molecular mechanisms of jSSc and highlight potential biomarkers for disease severity and progression.
To cite this abstract in AMA style:
Elbakri R, Robinson A, Sanyal A, Torok K. Whole Blood Transcriptome Profiling in Juvenile Systemic Sclerosis Patients Reveals Active Immune Upregulation and Enhanced Fibrotic Signature [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/whole-blood-transcriptome-profiling-in-juvenile-systemic-sclerosis-patients-reveals-active-immune-upregulation-and-enhanced-fibrotic-signature/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/whole-blood-transcriptome-profiling-in-juvenile-systemic-sclerosis-patients-reveals-active-immune-upregulation-and-enhanced-fibrotic-signature/