Session Information
Date: Tuesday, November 9, 2021
Title: Metabolic & Crystal Arthropathies – Basic & Clinical Science Poster II (1565–1583)
Session Type: Poster Session D
Session Time: 8:30AM-10:30AM
Background/Purpose: Gene expression studies of whole blood represent a powerful approach for understanding the pathogenesis of gout because differentially expressed transcripts may reflect the activation and response of immune cells to relevant and gout-specific stimuli. We have previously reported results (ACR 2019 abstract # 1232) of 48 statistically significant differentially expressed genes between gout cases (N=13) and non-gout controls (N =6). If the pathogenic mechanism that generates the differential expression has a genetic basis then we expect, 1) genetic variants near these genes to be associated with gout or its causal factor serum urate, and 2) that these variants co-localize with genetic variants that themselves associate with gene expression; i.e. the variants are cis-eQTLs.
Methods: We compared the genes whose expression was at least nominally associated with gout with results from a previously published transancestral meta analysis of serum urate and a separate GWAS of gout (Tin et al. 2019). We then used the Genotype-Tissue Expression (GTEX) database and COLOC (Giambartolomei et al. 2013) to verify whether the identified variants were associated with gene expression and colocalized with the serum urate signal.
Results: Only two out of 48 of the differentially expressed genes (GGT7 and FADS2) were found to harbor variants (cis-eQTLs-Figure 1), that co-localized with genome-wide significant loci for serum urate (but not with gout). These genes, GGT7 = gamma-glutamyltransferase 7, involved in glutathionine metabolism, and FADS2 = fatty acid desaturase 2, involved in fatty acid metabolism, were both underexpressed in gout cases relative to non-gout controls. These genes are therefore strong candidate genes for a causal pathogenic role in gout.
Conclusion: It may be the case that genetic variation for serum urate and gout that comprise eQTLs manifests as differentially expressed genes between gout cases and controls. However, the most frequent observation from our data is that no genetic signal was found that explained the differences. At least three factors may underly this observation: 1) It is possible that differentially expressed genes could be mediated through eQTLs acting in trans. 2) We lack statistical power to detect association of genetic variants and/or gene expression with gout. 3) Epigenetic regulation, and not genetic, may be the preeminent cause of the observed gene expression differences. As additional cell type-specific expression data and highly powered gout GWAS results become available we will continue to identify genetic loci that deserve attention as potentially pathogenetic features of hyperuricemia and gout.
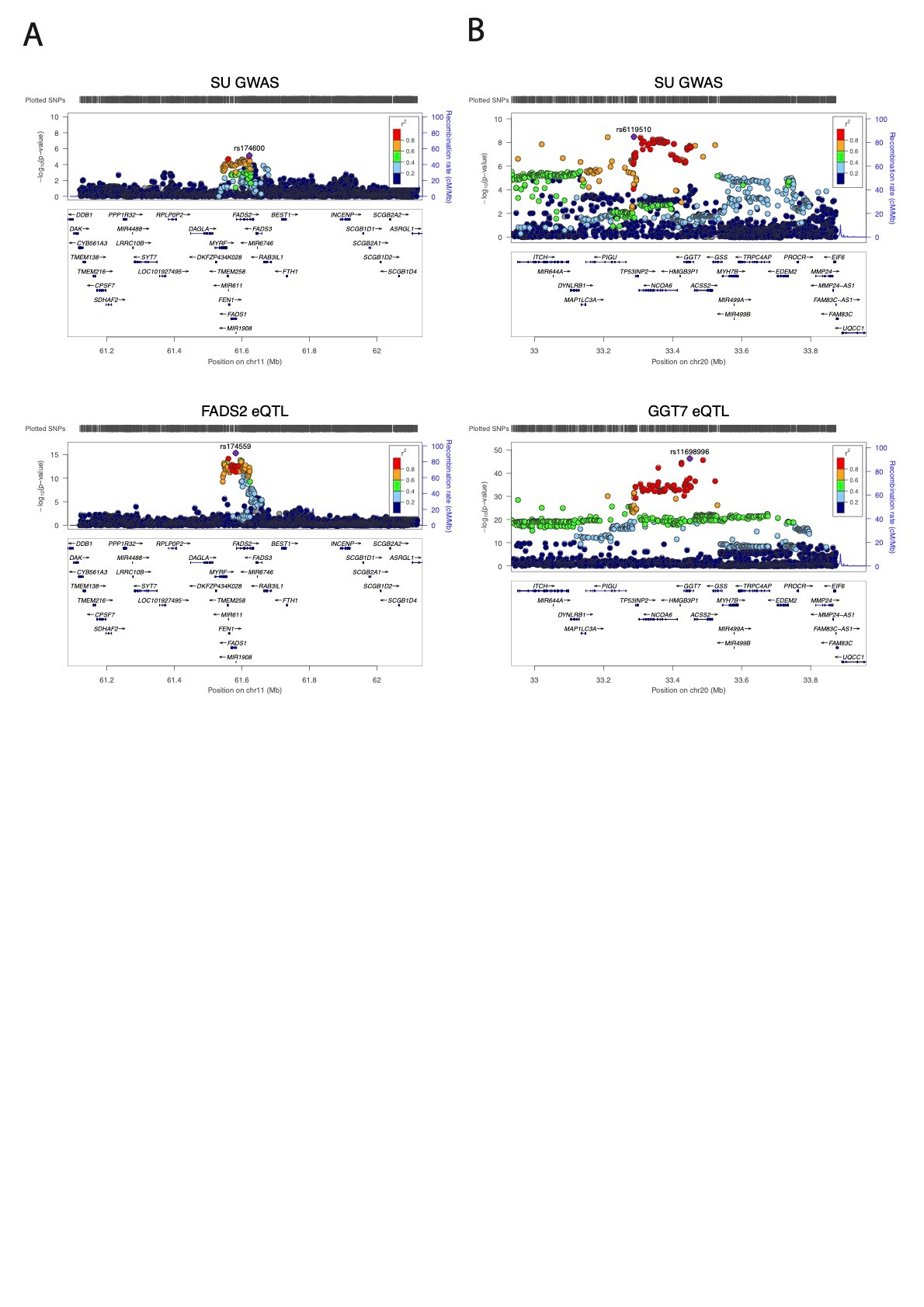 Figure 1. Colocalisation of eQTL and serum urate GWAS for FADS2 and GGT7. Regional association plots for serum urate (Tin et al. 2019) (EUR data) (top) and eQTL for (A) FADS2 (bottom left) and (B) GGT7 (bottom right). Dots indicate individual SNPs while position relative to the left y-axis indicates [-log10 (p-value)] of association. The blue line indicates the recombination rate across the locus. The lead serum urate and eQTL SNPs are indicated by a purple dot. The colouring of the surrounding SNPs indicates the strength of LD of the lead SNP according to the key on the left of each plot, measured as r2 found in the HapMap data for Europeans. The plot was generated using LocusZoom (Prium et al., 2010).
Figure 1. Colocalisation of eQTL and serum urate GWAS for FADS2 and GGT7. Regional association plots for serum urate (Tin et al. 2019) (EUR data) (top) and eQTL for (A) FADS2 (bottom left) and (B) GGT7 (bottom right). Dots indicate individual SNPs while position relative to the left y-axis indicates [-log10 (p-value)] of association. The blue line indicates the recombination rate across the locus. The lead serum urate and eQTL SNPs are indicated by a purple dot. The colouring of the surrounding SNPs indicates the strength of LD of the lead SNP according to the key on the left of each plot, measured as r2 found in the HapMap data for Europeans. The plot was generated using LocusZoom (Prium et al., 2010).
To cite this abstract in AMA style:
Reynolds R, Takei R, Edberg J, Sumpter N, Merriman T, Leask M. Whole Blood Gene Expression and eQTL Analysis Implicate GGT7 and FADS2 in Gout Pathogenesis [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/whole-blood-gene-expression-and-eqtl-analysis-implicate-ggt7-and-fads2-in-gout-pathogenesis/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/whole-blood-gene-expression-and-eqtl-analysis-implicate-ggt7-and-fads2-in-gout-pathogenesis/
