Session Information
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: Optimizing treatment of JIA continues to be a challenge. Biologic DMARD (bDMARD) therapies have significantly improved outcomes but is costly, may be more difficult to obtain, and can expose patients to risks of adverse effects. Importantly, many patients continue to have active disease. The ability to predict an individual’s likelihood of having worse JIA outcomes can inform treatment decisions. However, few strong clinical predictors of JIA outcomes have been identified and there remains a paucity of accurate prediction models. Here, we developed a machine learning approach to predict longitudinal outcomes in JIA.
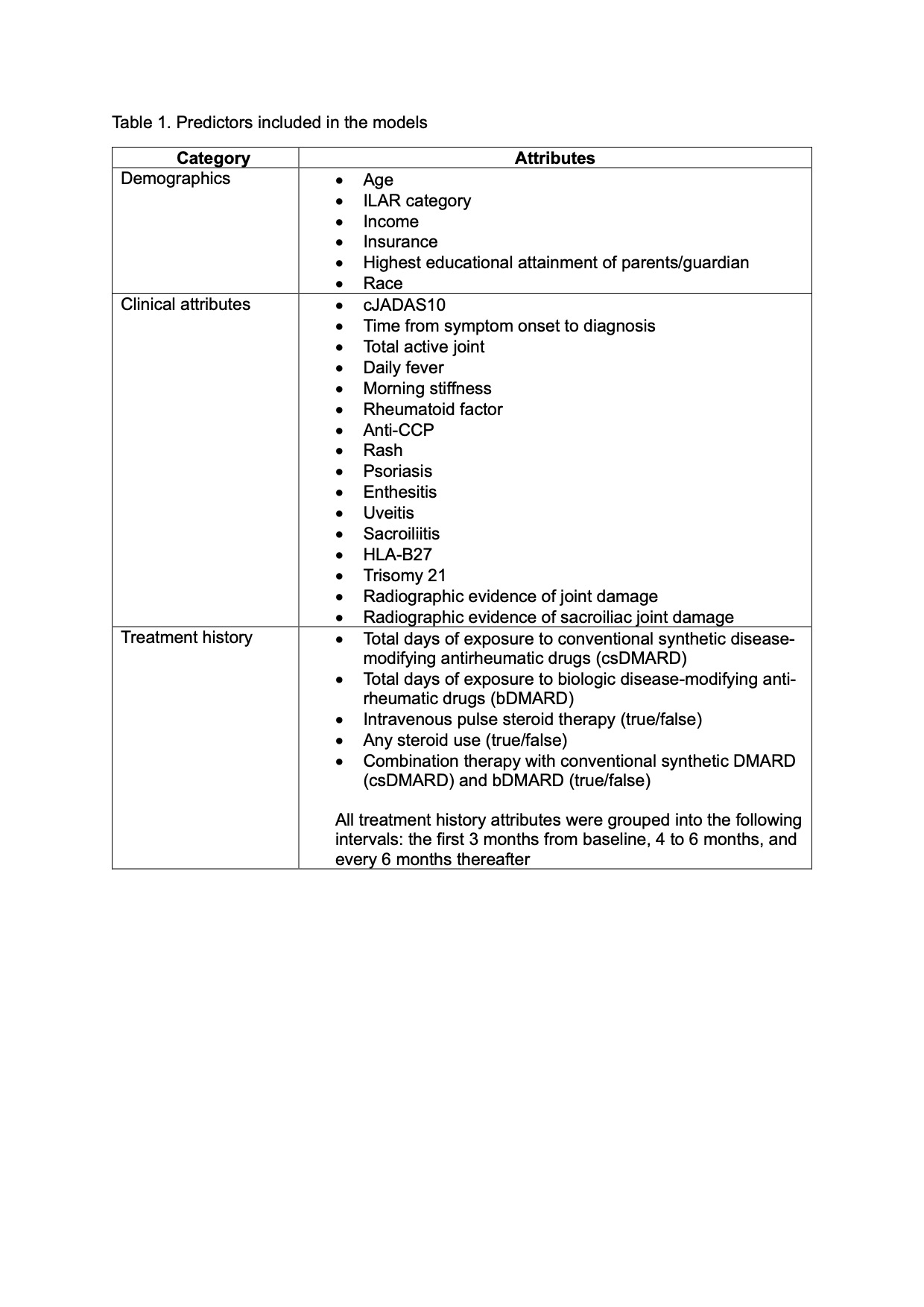
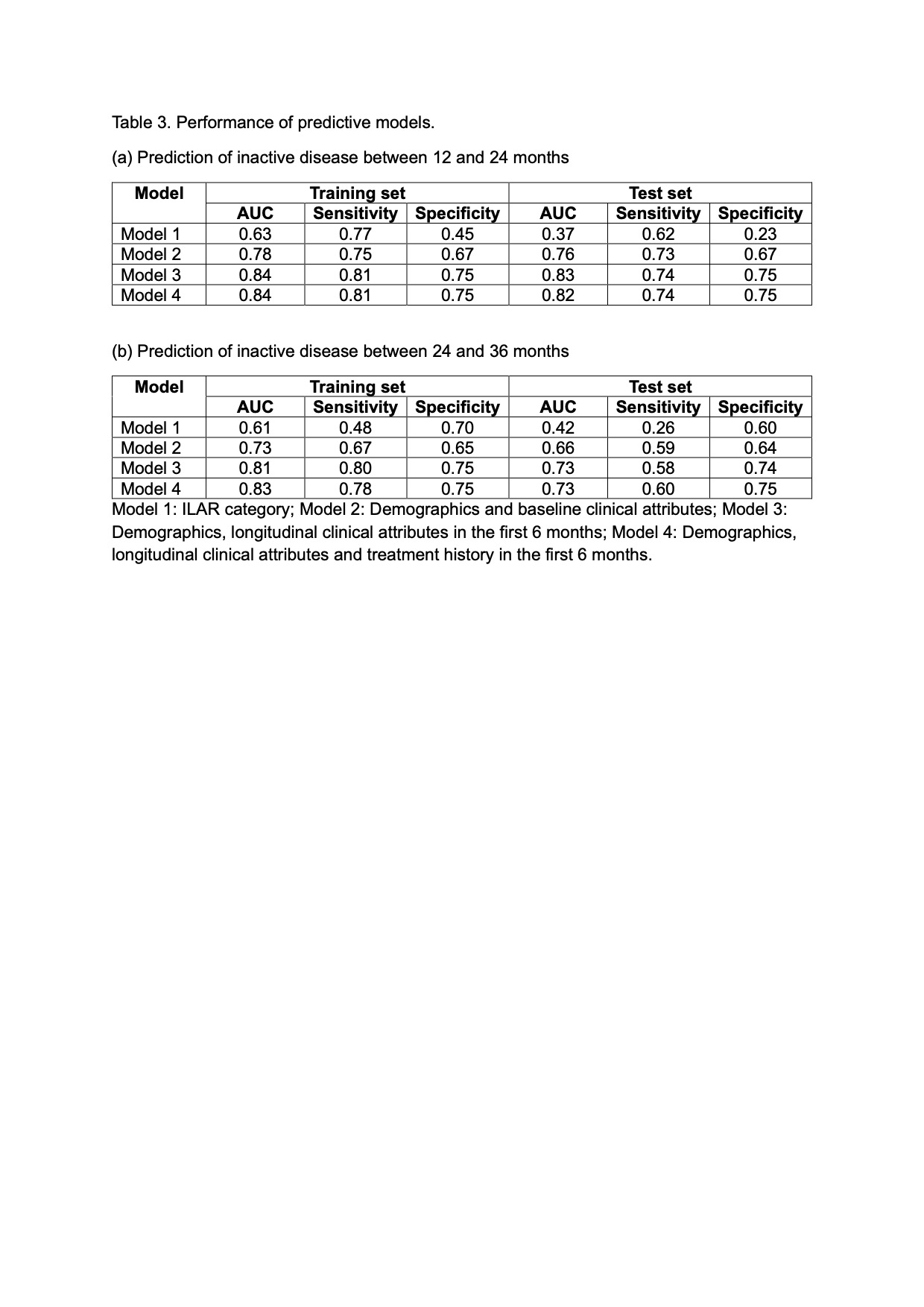
Methods: Retrospective cohort study of new onset JIA using the Childhood Arthritis and Rheumatology Alliance (CARRA) Registry (years 2015 to 2024). Lasso regression models were developed to predict achievement of inactive disease (cJADAS10 ≤2.5) at 2 time periods: (1) 12 and 24 months from baseline (initial registry visit); and (2) 24 and 36 months from baseline. The study dataset was randomly split into a training set (50%) for model development and a test set (50%) for model validation. We developed and compared separate models using the following predictors: (a) ILAR category – the strongest predictor of a non-remitting disease course reported in published studies (Model 1); (b) demographics and baseline clinical attributes (Model 2); (c) demographics, longitudinal clinical attributes in the first 6 months (Model 3); (d) demographics, longitudinal clinical attributes and treatment history in the first 6 months (Model 4). The area under the receiver operating curve (AUC) was used to select the most optimal models.
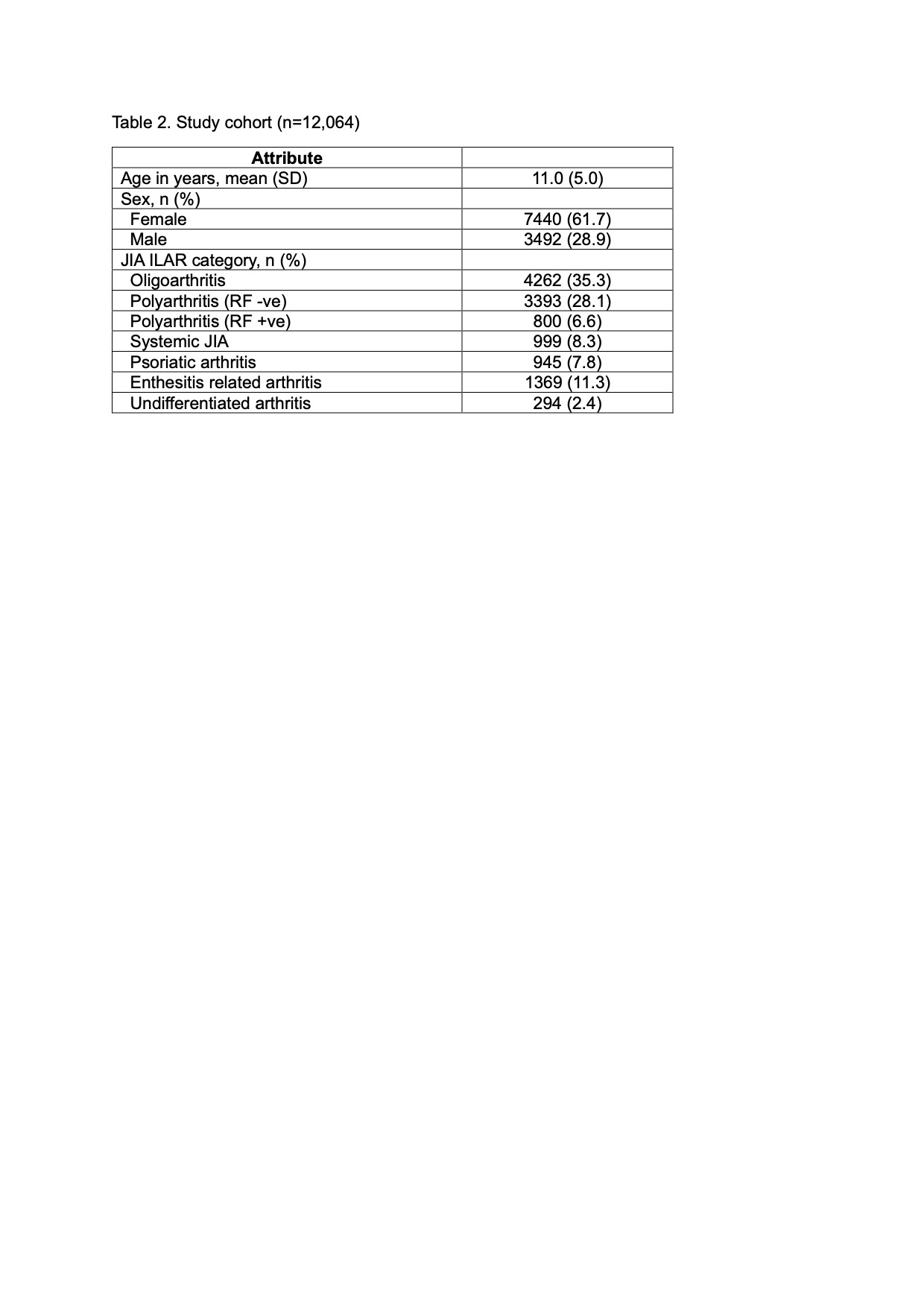
Results: A total of 12,064 JIA patients were included in the analysis (Table 2). When tested on independent test sets, models incorporating demographics and longitudinal clinical attributes best predicted inactive disease between 12 and 24 months (AUC: 0.83; sensitivity: 0.74; specificity: 0.75), and between 24 and 36 months (AUC: 0.73; sensitivity: 0.58; specificity: 0.74). The inclusion of treatment history in the prediction models achieved comparable results (Table 3). Models incorporating longitudinal clinical outcomes and/or treatment history outperformed models that included ILAR category alone, and those including demographics and baseline clinical attributes alone. Of note, prediction of inactive disease using ILAR category alone performed worse than chance on independent test sets (AUC < 0.5). cJADAS10 at baseline and at 6 months, age, and use of combination (bDMARD + csDMARD) therapy in the first 3 months best predicted inactive disease between 12 and 24 months. cJADAS10 at 6 months, total days of exposure to bDMARD in the first 3 months, and total joint count ≥5 at baseline were the most important attributes for predicting inactive disease between 24 and 36 months.
Conclusion: We demonstrated the feasibility of using machine learning models to predict longitudinal outcomes of JIA patients. Our models incorporated data collected during routine clinical visits. Incorporation of emerging sources of multi-omics data has the potential to improve the prediction of outcomes and inform treatment decision-making.
To cite this abstract in AMA style:
Ong M, Natter M, Schanberg L, Kimura Y. Using Machine Learning to Predict Inactive Disease in Juvenile Idiopathic Arthritis [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/using-machine-learning-to-predict-inactive-disease-in-juvenile-idiopathic-arthritis/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/using-machine-learning-to-predict-inactive-disease-in-juvenile-idiopathic-arthritis/