Session Information
Date: Monday, November 13, 2023
Title: (1264–1307) RA – Diagnosis, Manifestations, and Outcomes Poster II
Session Type: Poster Session B
Session Time: 9:00AM-11:00AM
Background/Purpose: Recent research has revealed a significant upregulation of IFNγ-related genes in RA patients, though the underlying mechanisms and implications are still unclear. To further investigate these gene expression alterations, we implemented single-cell RNA sequencing and performed an analysis on the peripheral blood mononuclear cells (PBMCs) of RA patients.
Methods: We executed a single-cell RNA sequencing study on peripheral blood samples from patients with anti-citrullinated protein antibody positive (ACPA+) and negative (ACPA-) RA. Cell subsets were identified, and their distribution and functional characteristics were compared based on ACPA presence.
Results: Our analysis identified 37,318 single cells grouped into 17 different cell types. We observed an expanded proportion of IL1B+ monocytes, IL7R+ T cells, and CD8+ CCL4+ T cells in ACPA+ RA patients. Enrichment analysis scores of interferon-gamma (IFNγ) response genes increased in nearly all monocytes and T cell subsets in ACPA+ RA. These patients exhibited heightened interactions between IFNγ and IFNγ receptors upstream of IFNγ response genes. Cell-cell interaction was significantly amplified between monocytes and T cells in ACPA+ RA. Additionally, we discovered upregulated IFNγ-driven transcription factors (TFs) such as STATs and IRFs in ACPA+ RA. Major target genes of STATs and IRFs were identified, with IFITM3 and IFITM2 standing out as strongly associated target genes. A significant positive correlation was also found between IFITM2, IFITM3, and elevated serum levels of IFNγ, a signature cytokine of Th1 cells. This finding supports their potential clinical implications in ACPA+ RA.
Conclusion: Our findings offer novel insights emphasizing the critical role of IFNγ in regulating Th1 skewing in the pathogenesis of ACPA+ RA, as compared to ACPA- RA. The increased interactions between monocytes and T cells and high serum levels of IFNγ, particularly in ACPA+ RA, provide a more comprehensive understanding of the pathogenesis of ACPA+ RA.
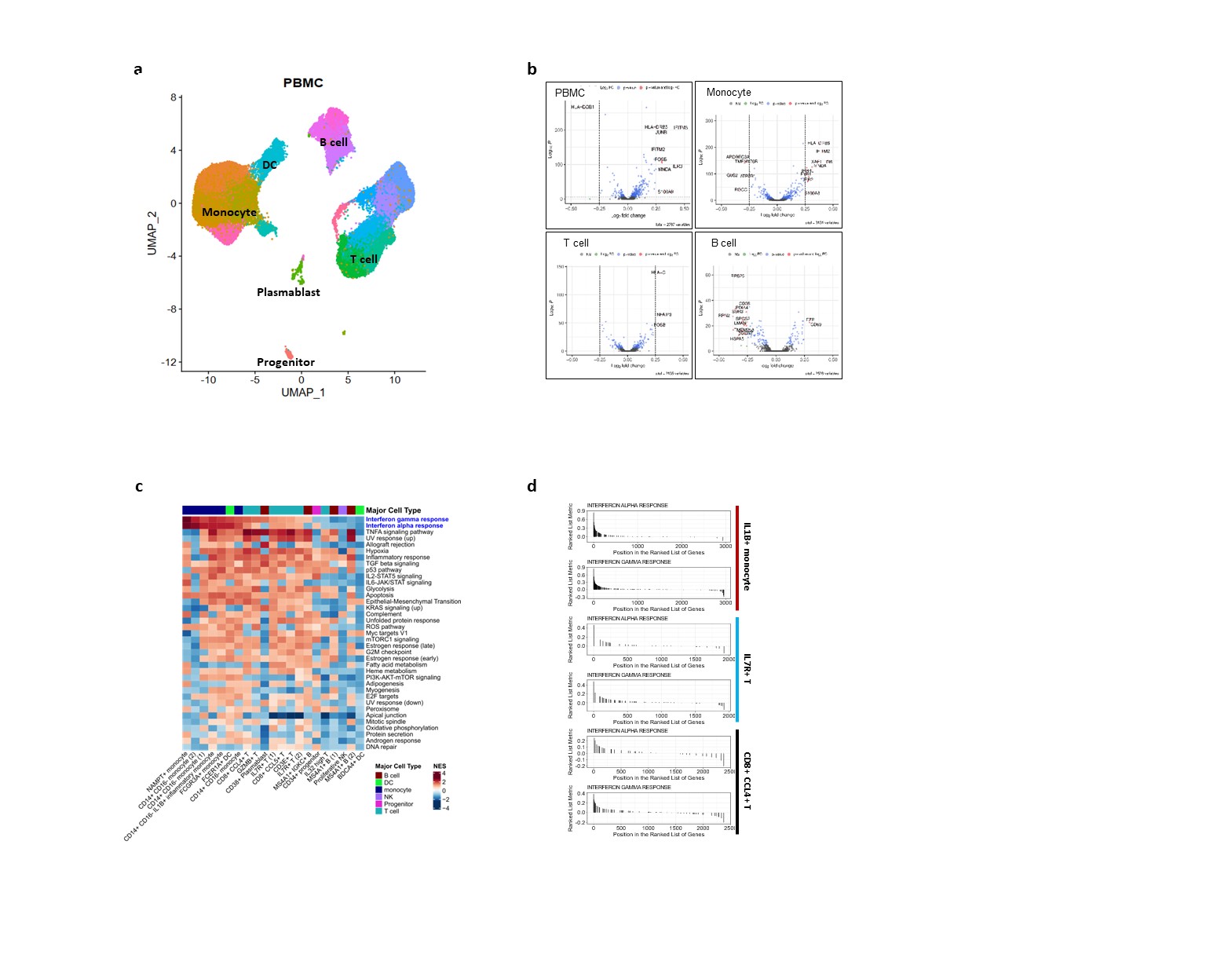
(a) t-SNE map based on single-cell RNA-seq data obtained from PBMC of 16 rheumatoid arthritis (RA) patients, with or without anti-CCP antibody, showing 17 different cell types after clustering with 37,318 cells. (b) Volcano plot for PBMC, T, B, and monocyte, comparing ACPA+ vs. ACPA- groups, showing differentially expressed genes obtained by comparison. (c) The heatmap displays the results of gene set enrichment analysis for genes whose expression levels change in ACPA-compared to ACPA+ for 17 cell types. Column annotation of heatmap showing the major cell types (T, B, DC, monocyte, NK, progenitor) indicated for each cell type. (d) The GSEA graph shows the results of IFN-alpha and IFN-gamma response gene sets for three types of subcellular groups that increase in ACPA+ patient groups.
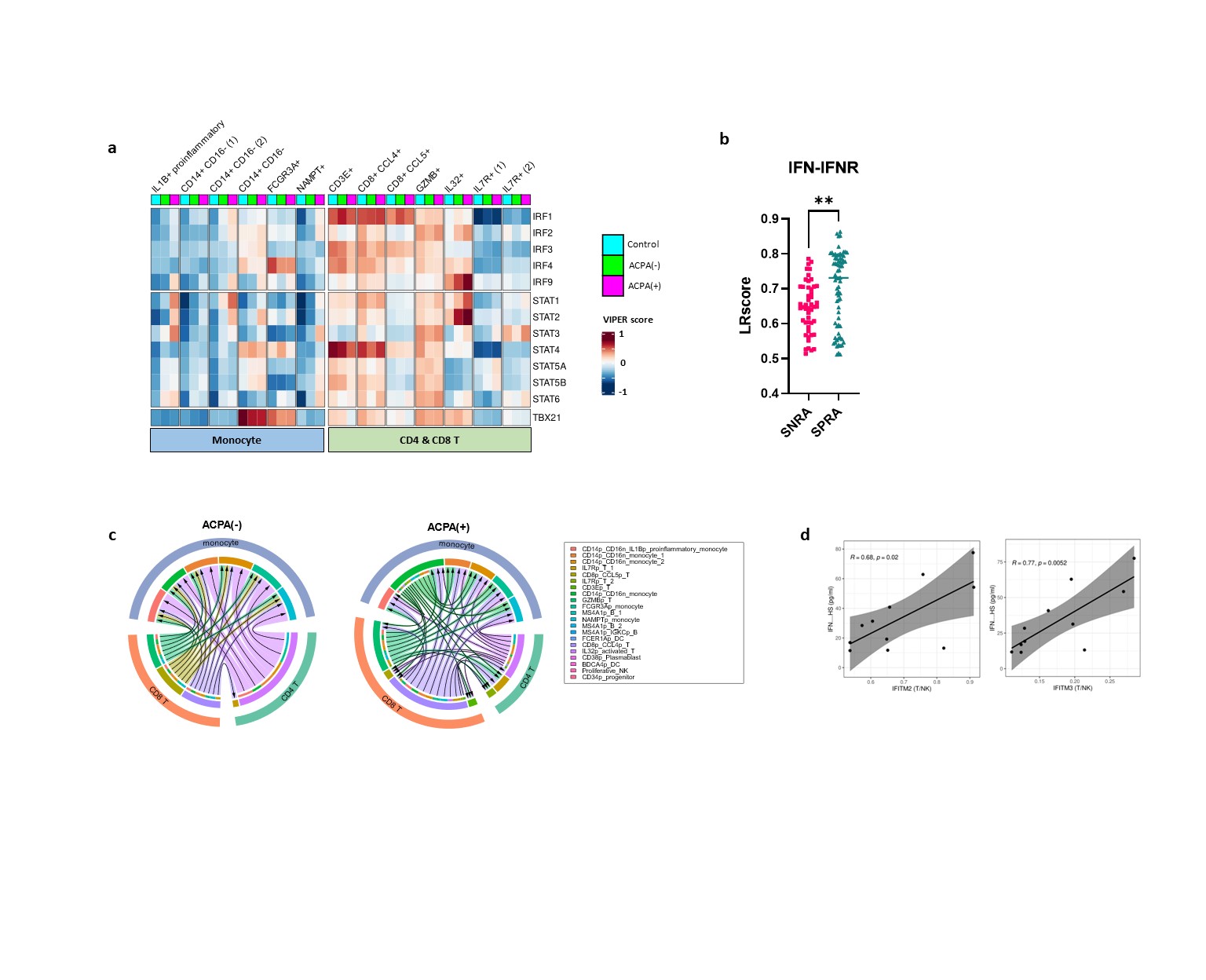
(a) Heatmap representing the activity score of STAT and IRF families predicted to regulate the expression of IFITM2 and IFITM3. (b) The bar graph represents the LR score for IFN-IFNR in the ligand-receptor analysis results. (c) Chord diagram showing the difference in the interaction of IFN-IFNR between monocyte-T cells in ACPA- and ACPA+ groups. The legend on the left side of the diagram represents the subcellular types of monocyte and T cells seen in the second layer of the diagram. (d) Graph showing the correlation between the degree of mRNA expression of IFITM2/3 for each patient and Interferon concentration measured in serum.
To cite this abstract in AMA style:
Hong B, You S, Lee N, Kim J, Lee K, Ju J, Kim W, Kim H. Upregulation of IFNγ-Response Genes in Monocytes and T Cells Identified by Single-Cell Transcriptomics in Patients with Anti-CCP Antibody-Positive Rheumatoid Arthritis (RA) [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/upregulation-of-ifn%ce%b3-response-genes-in-monocytes-and-t-cells-identified-by-single-cell-transcriptomics-in-patients-with-anti-ccp-antibody-positive-rheumatoid-arthritis-ra/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/upregulation-of-ifn%ce%b3-response-genes-in-monocytes-and-t-cells-identified-by-single-cell-transcriptomics-in-patients-with-anti-ccp-antibody-positive-rheumatoid-arthritis-ra/