Session Information
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: Antiphospholipid syndrome (APS) is a complex disease characterized by the presence of anti-phospholipid antibodies (aPLs). Its significant clinical heterogeneity brings clinical and therapeutical challenges. To date few studies have focused on APS subtype-specific stratification, which could be important for clinical management and prognostic evaluation in APS. We therefore aimed to study whether unrecognized disease subgroups can be identified by developing a novel integrated metrics for unsupervised clustering algorithm.
Methods: A two-stage (training and validation) study was conducted. Totally 808 APS patients were enrolled (training cohort: n=442, validation cohort: n=366). Of which, the validation cohort was consisted of an internal sub-cohort (n=245) and an external sub-cohort (n=121). An unsupervised K-means clustering algorithm was applied to stratify the study subjects, according to multiple indicators, e.g. age, gender, obstetric complications, thrombotic events, non-criteria APS manifestations, immunoglobulin and complement levels, etc. The different clusters were then evaluated for their prognostic outcomes. Finally, we conducted a proteomic analysis for primary APS patients derived from three Clusters (n=25) and healthy controls (n=9).
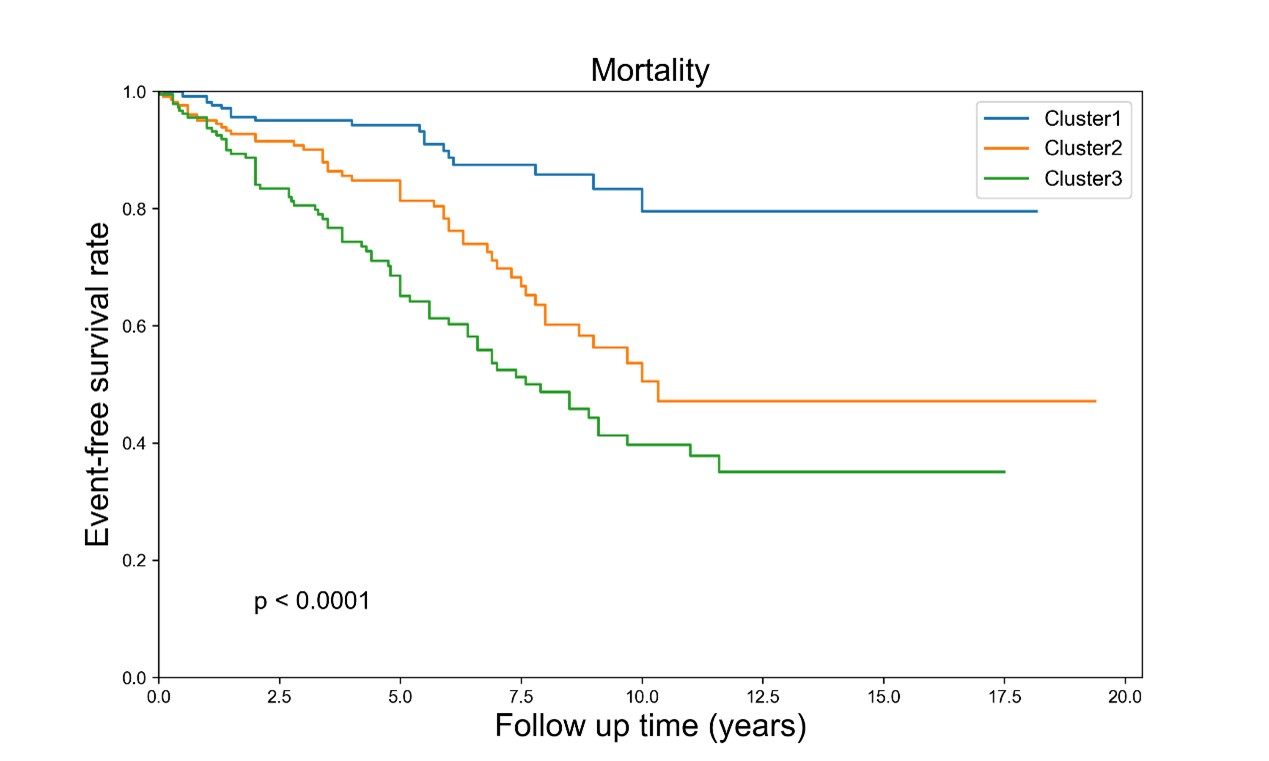
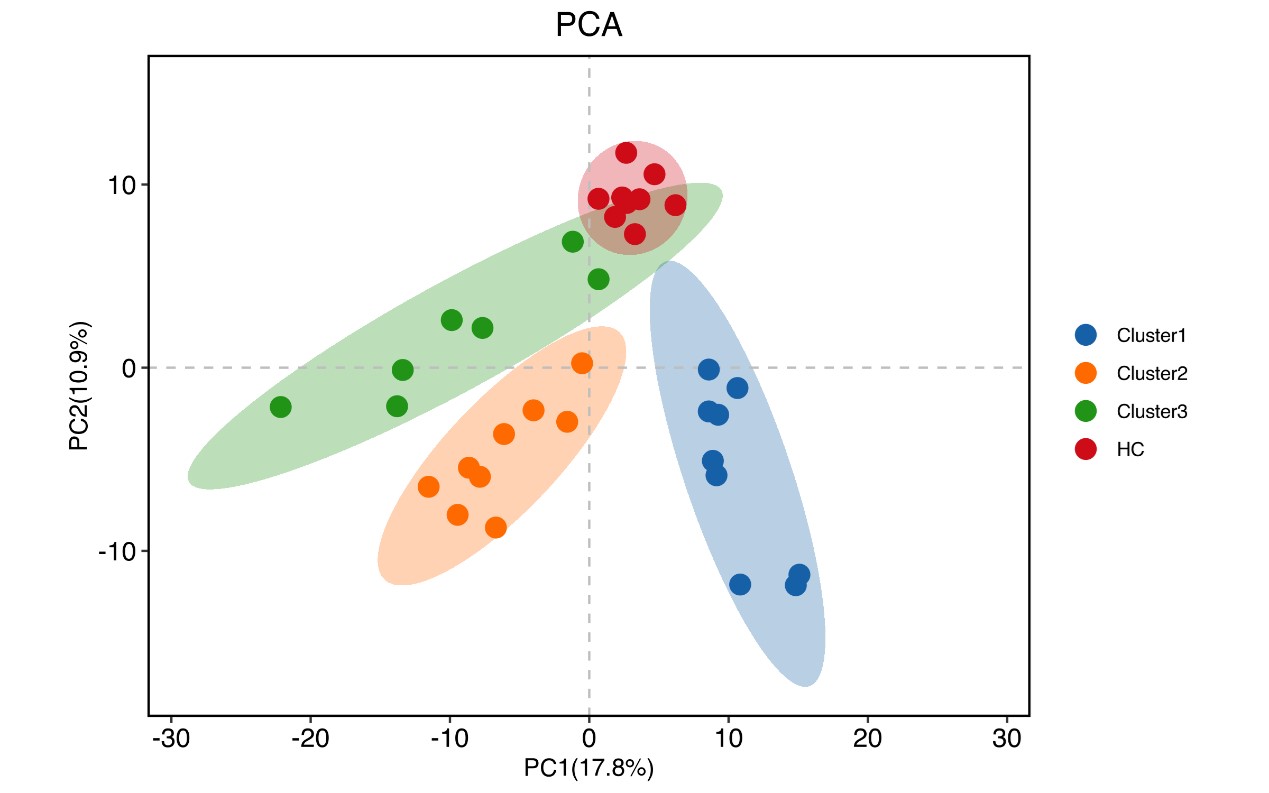
Results: By using the unsupervised clustering algorithm, we identified three distinctive clusters. Cluster 1 was predominantly consisted of female patients with obstetric complications and very few patients were with thrombotic APS. These clustered patients showed a good prognosis. Cluster 2 was also mainly consisted of female patients, but most of them suffered from thrombosis. These clustered patients displayed apparent non-criteria APS manifestations and/or metabolic syndromes, and an intermediate prognosis. These patients also had a significant decrease in complement C3 and C4. In contrast, Cluster 3 was dominantly consisted of male patients, and most of them had thrombotic APS, with a high proportion of primary APS. The clustered patients displayed a poor prognosis (shown in Figure 1). Furthermore, positivity of the triple aPLs were gradually increased from Cluster 1 to Cluster 3. Proteomic analysis identified 157 significant differentially expressed molecules across three clusters, mainly involving in complement and coagulation cascades. Principal component analysis (PCA) of proteomics clearly discriminated APS patients from HCs, as well as the patients derived from three Clusters (shown in Figure 2).
Conclusion: By the unsupervised machine learning approach, we identified three distinct APS subgroups. Each subgroup displayed unique clinical manifestations and molecular features, as well as a marked difference in prognostic outcomes. Our novel findings will provide new insight into the pathogenesis of APS, and may also provide new clues for clinical management and prognostic evaluation in APS.
To cite this abstract in AMA style:
Chen C, Zhang A, Cheng J, Yao Z, Meng J, Qin Y, Lu Q, Li Y, Liu X, Li T, Hou C, Tang Y, Liu H, Xu N, Dong S, Li X, Xu F, Guo J, Li C. Unsupervised Machine Learning Improves Clinical Stratification and Prognostic Evaluation for Antiphospholipid Syndrome: A Large Cohort Study from China [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/unsupervised-machine-learning-improves-clinical-stratification-and-prognostic-evaluation-for-antiphospholipid-syndrome-a-large-cohort-study-from-china/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/unsupervised-machine-learning-improves-clinical-stratification-and-prognostic-evaluation-for-antiphospholipid-syndrome-a-large-cohort-study-from-china/