Session Information
Date: Monday, November 13, 2023
Title: (0934–0964) Systemic Sclerosis & Related Disorders – Basic Science Poster
Session Type: Poster Session B
Session Time: 9:00AM-11:00AM
Background/Purpose: Systemic sclerosis (SSc) is an autoimmune disease characterized by skin and internal organ fibrosis, vascular abnormalities, and immunological disturbances. Monocytes play a critical role in SSc, but their contribution to disease pathogenesis remains unclear. A better understanding of gene expression changes in SSc monocytes and their associations with disease complications may lead to novel therapies.
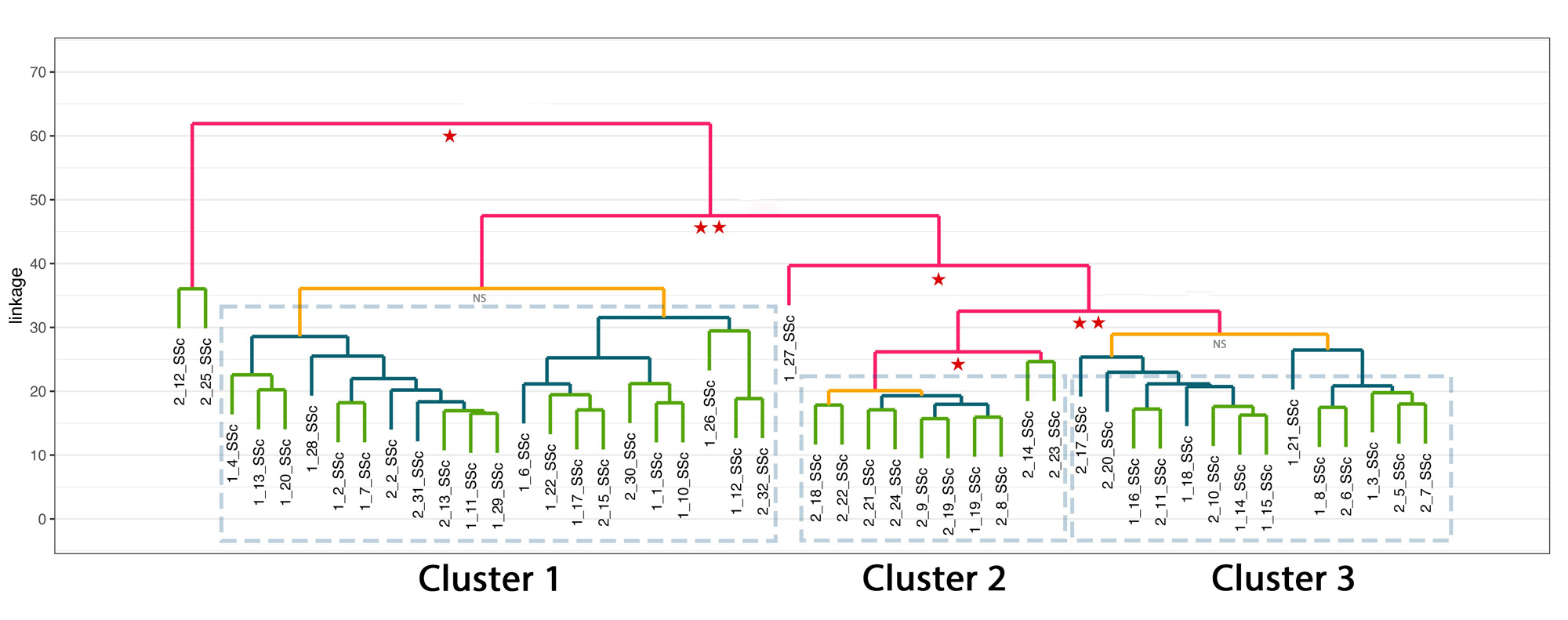
Methods: Peripheral blood mononuclear cells were obtained from 48 SSc patients and 15 controls (HC) after informed consent and under an institutional approved IRB protocol. Magnetic bead separation was used to isolate monocytes and bulk RNA sequencing performed using the Illumina NextSeq platform. DESeq2 was used to identify differentially expressed genes between groups. We employed hierarchical clustering to group samples exhibiting similar transcriptional trends. Functional enrichment analysis was performed using ORA with the GO database to investigate the biological processes linked to each group. Pearson correlation was used for mRSS. DEG, GO and correlation analysis were conducted using R version 4.3.0. Pathways analysis for clinical features was performed using Ingenuity Pathway Analysis.
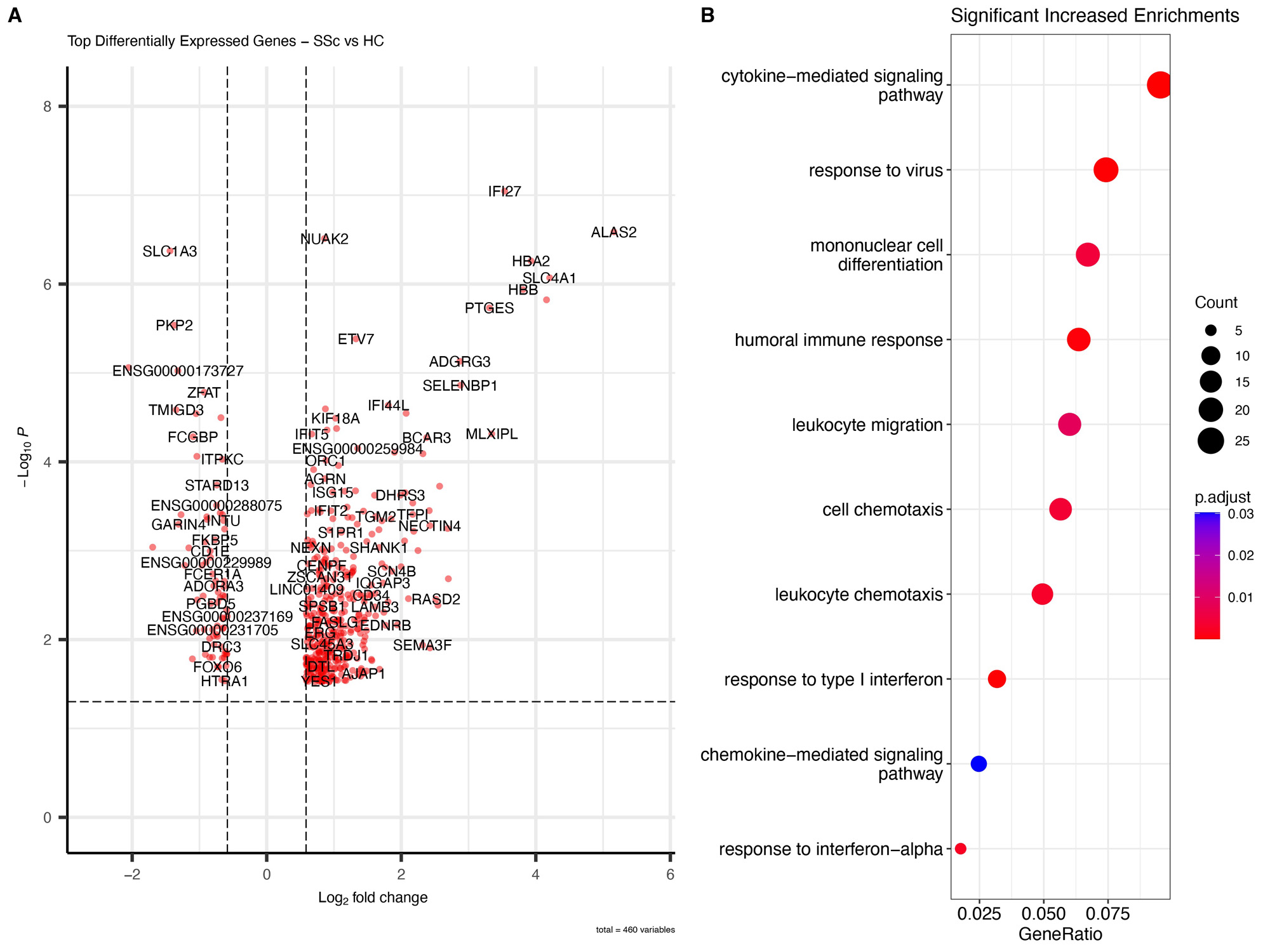
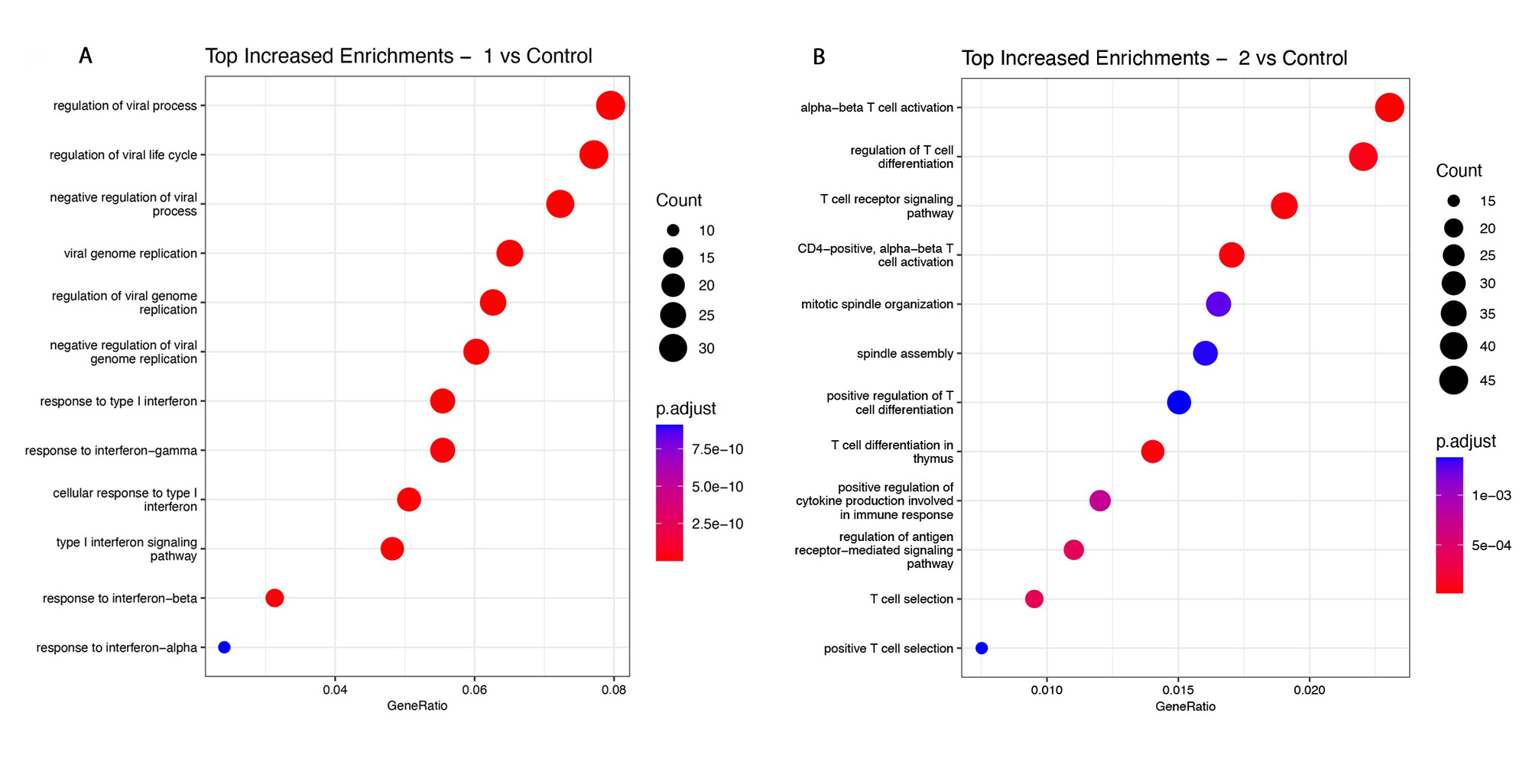
Results: 460 genes were found to be differentially regulated between SSc and HC. Interferon-related genes (IFI27, IFIT5, IFI44L, SIGLEC) were among top differentially expressed, along with genes not previously linked to SSc, including NUAK2, a TGFb induced protein kinase belonging to the AMPK family (FC 1.82, padj 0.001), and LGALS3BP, a secreted galectin-3 binding protein, that regulates cell-cell interactions and inflammation (FC 2.08, padj 0.04). Hierarchical clustering in SSc unveiled three groups. Cluster 1 (“inflammatory-like”, 21 patients) predominantly displayed interferon related pathways, and included >80% of SSc patients with pulmonary hypertension (PH) from the entire SSc cohort. Cluster 2 (“T cell-like”, 10 patients) exhibited enrichment in multiple pathways associated with T cell activation and contained the majority of patients undergoing immunosuppressive therapy. Cluster 3 (“normal-like”, 14) closely resembled the control group. Three patients did not belong to any clusters. There were significant associations between gene expression and clinical features. Interferon signaling pathway was the top most upregulated pathway in SSc-PH patients versus HC, but did not differentiate patients with and without PH. Cardiomyopathy patients had enhanced FAK and integrin signaling, and phagosome formation pathways. IPA analysis of SSc-ILD vs HC revealed association with pulmonary fibrosis signaling pathway with increased expression of SERPINE1 and PLAU genes. mRSS was highly correlated with XPR1, a novel gene implicated in phosphate homeostasis (r=0.72, padj 0.006).
Conclusion: Our results suggest that SSc patients can be classified into distinct groups based on transcriptomic profiles of monocytes, which correlate with specific clinical characteristics. The DEGs for clinical features revealed significant connections to the disease manifestations, potentially opening the avenue for new therapeutic targets.
To cite this abstract in AMA style:
Dinc M, El Adili F, Lui J, Ligresti G, Lafyatis R, Trojanowska M, Bujor A. Transcriptomic Analysis of Scleroderma Monocytes Identifies Distinct Clinically Relevant Clusters and Novel Genes Associated with Disease Complications [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/transcriptomic-analysis-of-scleroderma-monocytes-identifies-distinct-clinically-relevant-clusters-and-novel-genes-associated-with-disease-complications/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/transcriptomic-analysis-of-scleroderma-monocytes-identifies-distinct-clinically-relevant-clusters-and-novel-genes-associated-with-disease-complications/