Session Information
Session Type: ACR Abstract Session
Session Time: 2:30PM-4:00PM
Background/Purpose: Lupus nephritis (LN) complicates up to 60% of patients with systemic lupus erythematosus (SLE) and carries a high morbidity and mortality. The definitive diagnosis is based on kidney biopsy. This is invasive and not always readily available, thus delaying treatment. Sometimes multiple biopsies are required over the course of the disease. Importantly, while renal pathology is accurate at describing the morphology of renal disease, the underlying biology and molecular pathways are not thoroughly assessed. Urine proteomics is a non-invasive strategy that may provide insights regarding ongoing renal disease.
Methods: One thousand proteins were quantified (RayBiotech Kiloplex assay) on a total of 112 longitudinal urine samples from 32 SLE patients with active LN and 7 healthy controls (HC) enrolled in the Accelerating Medicines Partnership (AMP). All patients underwent treatment as directed by their own physicians. Differentially excreted proteins at baseline (SLE vs HC, proliferative vs membranous LN, responders vs non responders) were identified using a linear model with moderated t statistic. Response to treatment was defined based on proteinuria at 1 year as “complete” (< 0.5g/24h) or “partial” (50% reduction but >0.5/24h). In the longitudinal analysis, a mixed model was employed to identify markers associated with proteinuria. Pathway enrichment analysis was performed using the genes coding for the differentially excreted analytes using Ingenuity Pathway Analysis (IPA) and other publicly available pathway libraries.
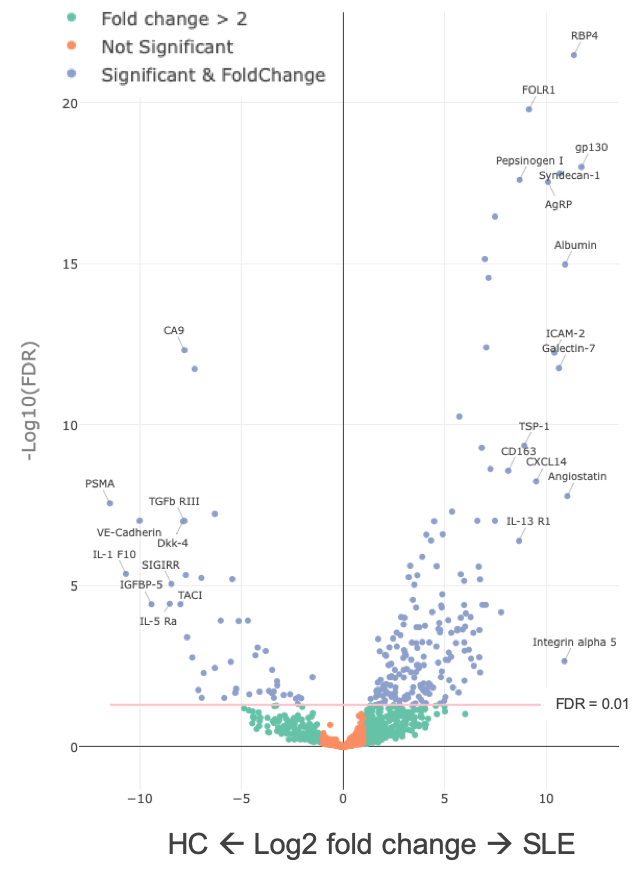
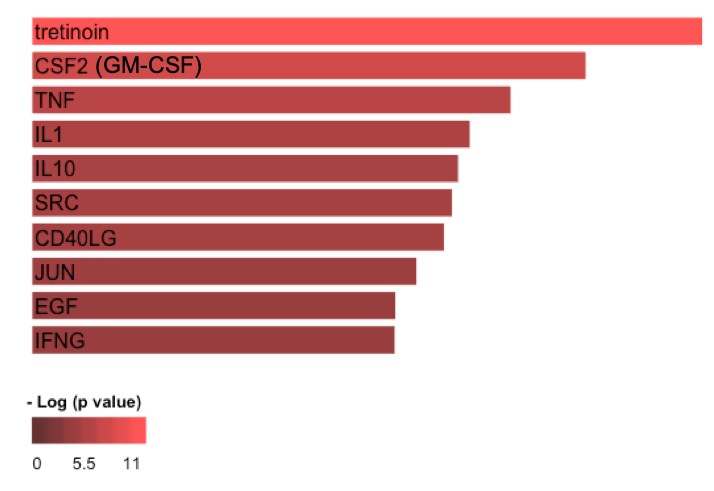
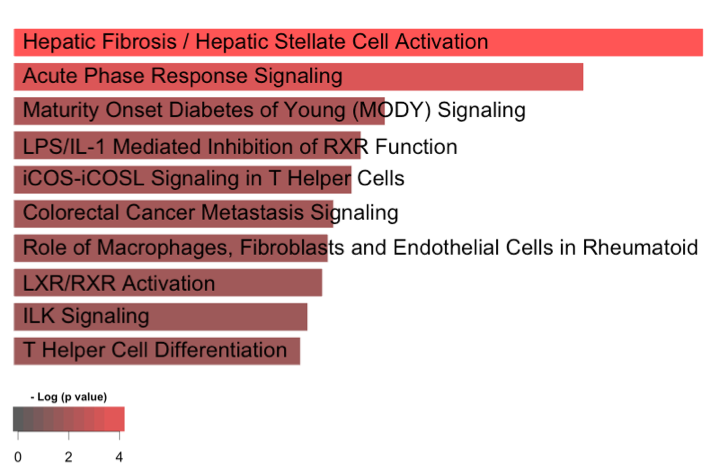
Results: There were 186 proteins increased in SLE patients (Fig. 1). The most enriched pathway was TNFa (p< 0.001). We found 74 differentially excreted proteins comparing proliferative and pure membranous LN. CD4, MCP-1, MIP-1a, RANTES, IL-16, and IL-7, markers involved in CD4 T cell and monocyte biology, were enriched in proliferative disease. A few targets were exclusively identified in either class (i.e. CD4 in proliferative nephritis). We used a longitudinal model to identify specific urine proteins associated with worse proteinuria as a marker of severity. Proteinuria was associated with 105 markers (FDR < 0.05), the strongest association being CD163 (p = 10-9), a phagocyte marker. IPA implicated several pathways involving fibrosis, acute phase response, LPS/IL1, RXR, ICOS signaling and macrophage/fibroblasts (Fig. 2). Next, we identified 27 differentially excreted proteins in non-responders. IPA revealed that tretinoin, GM-CSF, TNF, and IL1 were among the top upstream regulators (Fig. 3).
Conclusion: There is an inflammatory signature in the urine of patients with LN implicating monocyte and TNFa pathways. These signatures are associated with proliferative disease, worse proteinuria, and non-response to treatment. Of note, TNFa is involved in LN and has therapeutic potential. In phase 1 of AMP, monocytes were the main urine cell type identified by singe cell RNA sequencing in patients with LN. These results suggest that urine proteomics might identify and infer active pathological mechanisms in LN, paving the way for a more personalized approach to treatment. Further work in Phase 2 of AMP is being pursued to validate and extend these findings.
To cite this abstract in AMA style:
Fava A, Zhang Y, Buyon J, Belmont H, Izmirly P, Mohan C, Zhang T, Accelerating Medicines Partnership T, Petri M. Toward a Liquid Biopsy for Lupus Nephritis: Urine Proteomic Analysis of SLE Identifies Inflammatory and Macrophage Signatures [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/toward-a-liquid-biopsy-for-lupus-nephritis-urine-proteomic-analysis-of-sle-identifies-inflammatory-and-macrophage-signatures/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/toward-a-liquid-biopsy-for-lupus-nephritis-urine-proteomic-analysis-of-sle-identifies-inflammatory-and-macrophage-signatures/