Session Information
Date: Monday, November 11, 2019
Title: Systemic Sclerosis & Related Disorders – Basic Science Poster
Session Type: Poster Session (Monday)
Session Time: 9:00AM-11:00AM
Background/Purpose: Systemic Sclerosis (SSc) is an autoimmune disease characterized by fibrosis and inflammation. Multiple organ systems are affected including the skin, gastrointestinal tract, vasculature, and lungs (Katsumoto, Whitfield, & Connolly, 2011). Gene expression analyses have identified four distinct molecular subtypes (inflammatory, fibroproliferative, normal-like, and limited) despite the heterogenous presentation of the disease. Microbial dysbiosis has been implicated previously in SSc skin and lower GI tract, (Arron et al., 2014; Volkmann, 2017), but no esophageal characterization has been completed. This study characterizes the esophageal microbiome of SSc patients and explores patterns in microbial profiles.
Methods: RNA sequencing was completed on 20 SSc patient and 3 healthy control samples. Sequencing reads were run through Integrated Metagenomic Sequence Analysis (IMSA) to extract non-human microbial reads remaining after initial BLAST to the h19 NCBI human genome. Remaining reads were BLAST to the NCBI nt nucleotide database and taxonomic information was acquired using NCBI taxonomy lookup tool. Non-spurious hits were removed, and samples were rarefied to the lowest read count. Metrics such as the alpha diversity and Firmicutes/Bacteroidetes Ratio, which is associated with gut inflammatory diseases, were calculated. Partition Around Medoids (PAM) clustering based on k clusters was conducted on metagenomic features to group individuals into microbiome enterotypes.
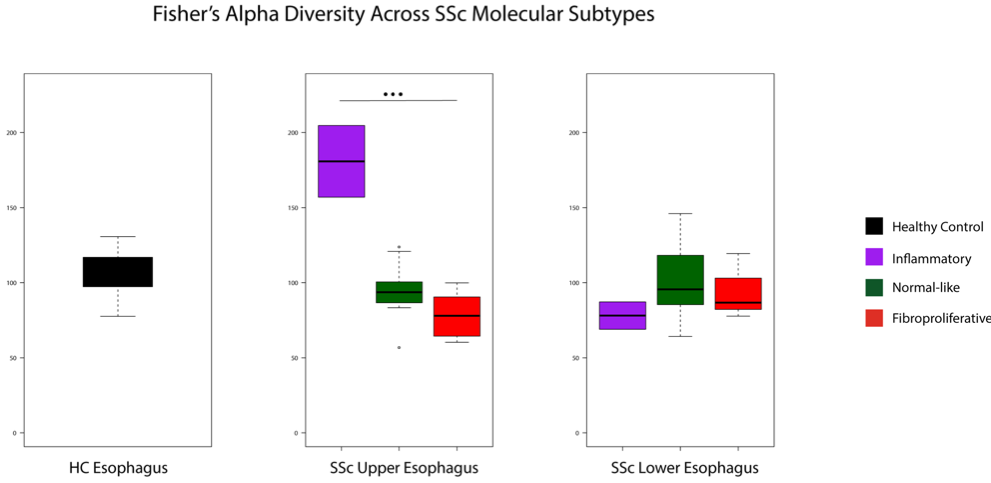
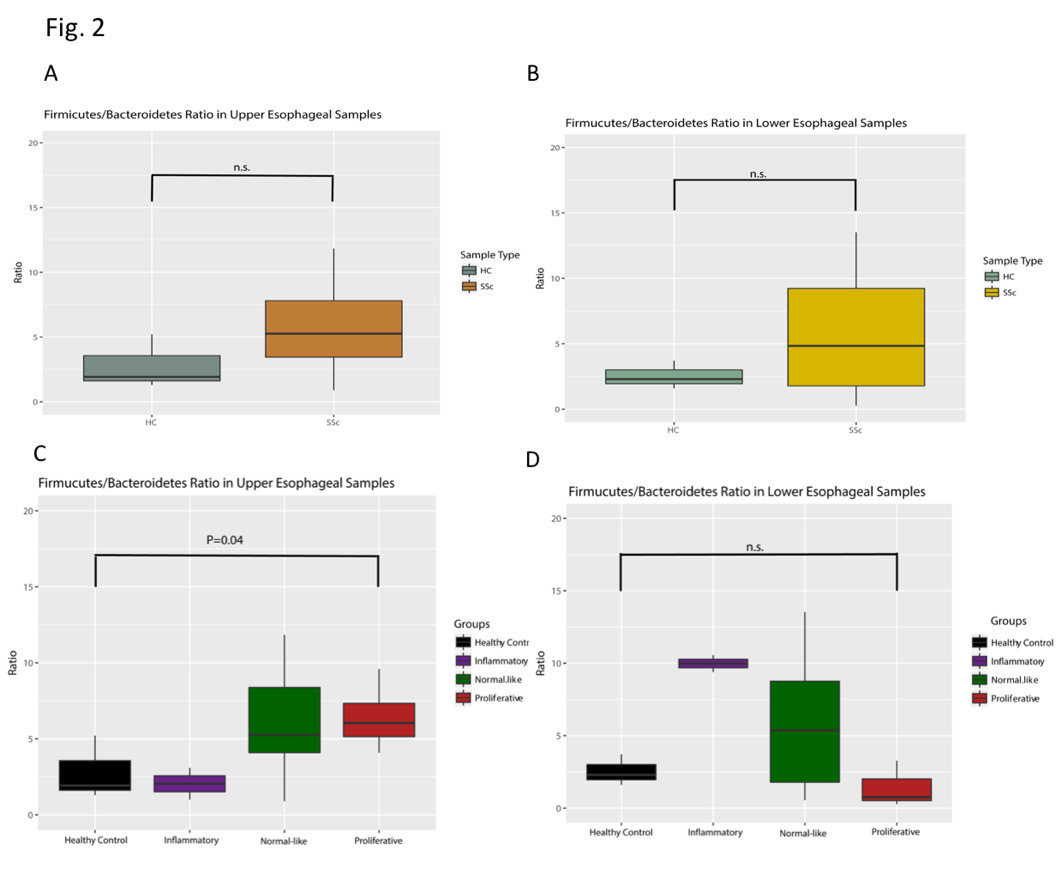
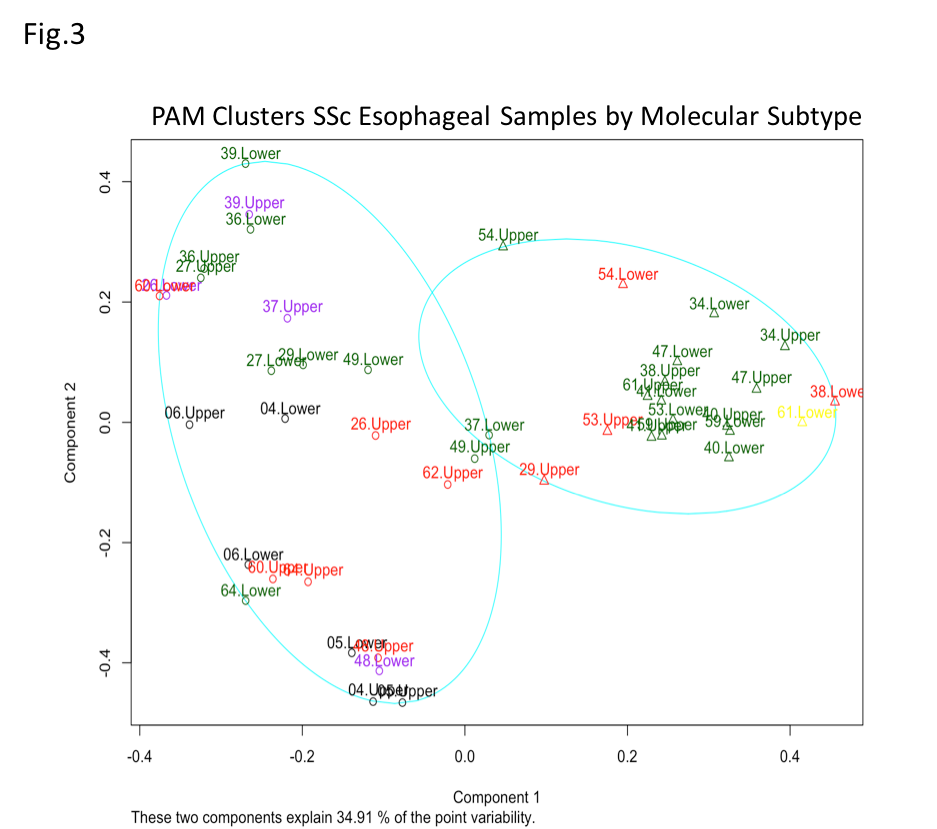
Results: The alpha diversity of SSc samples was lower than that of control samples across the upper and lower esophagus (Wilcoxon Signed-Rank Test, n.s). When parsed by esophageal site and molecular subtype, the alpha diversity was higher in inflammatory samples of the upper esophagus (Kruskal-Wallis Test, p< 0.05) and slightly elevated in normal-like and fibroproliferative samples of the lower esophagus (Fig. 1). Concurrent with differences in alpha Diversity, the Firmicutes to Bacteroidetes ratio was elevated in patient samples when compared to healthy controls (Fig. 2A and 2B). When stratified by SSc molecular subtype, Firmicutes/Bacteroidetes ratios were higher in SSc upper esophageal samples which call to the normal-like and fibroproliferative subtypes, and higher in SSc lower esophageal inflammatory and normal-like samples (Fig. 2C and 2D; Kruskal-Wallis test p< 0.05 and n.s., respectively). Samples clustered to two groups irrespective of site, and there is a significant association between molecular subtype and cluster membership (Fig. 3; Fisher’s Exact Test, p< 0.05).
Conclusion: There are patterns and differences in microbial diversity and membership that distinguish SSc and healthy controls, and SSc samples across molecular type. Several of these associations are statistically significant and affirm a potential association between the microbiome and SSc pathogenesis. Work investigating specific microbial membership and gene expression associations are motivated by these findings.
To cite this abstract in AMA style:
Espinoza M, Mehta B, Wang Y, Hoffman A, Aren K, Carns M, Kosarek N, Wood T, Hinchcliff M, Whitfield M. The Diversity and Community Metrics of the Esophageal Microbiome of SSc Patients [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/the-diversity-and-community-metrics-of-the-esophageal-microbiome-of-ssc-patients/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/the-diversity-and-community-metrics-of-the-esophageal-microbiome-of-ssc-patients/