Session Information
Session Type: Poster Session A
Session Time: 9:00AM-11:00AM
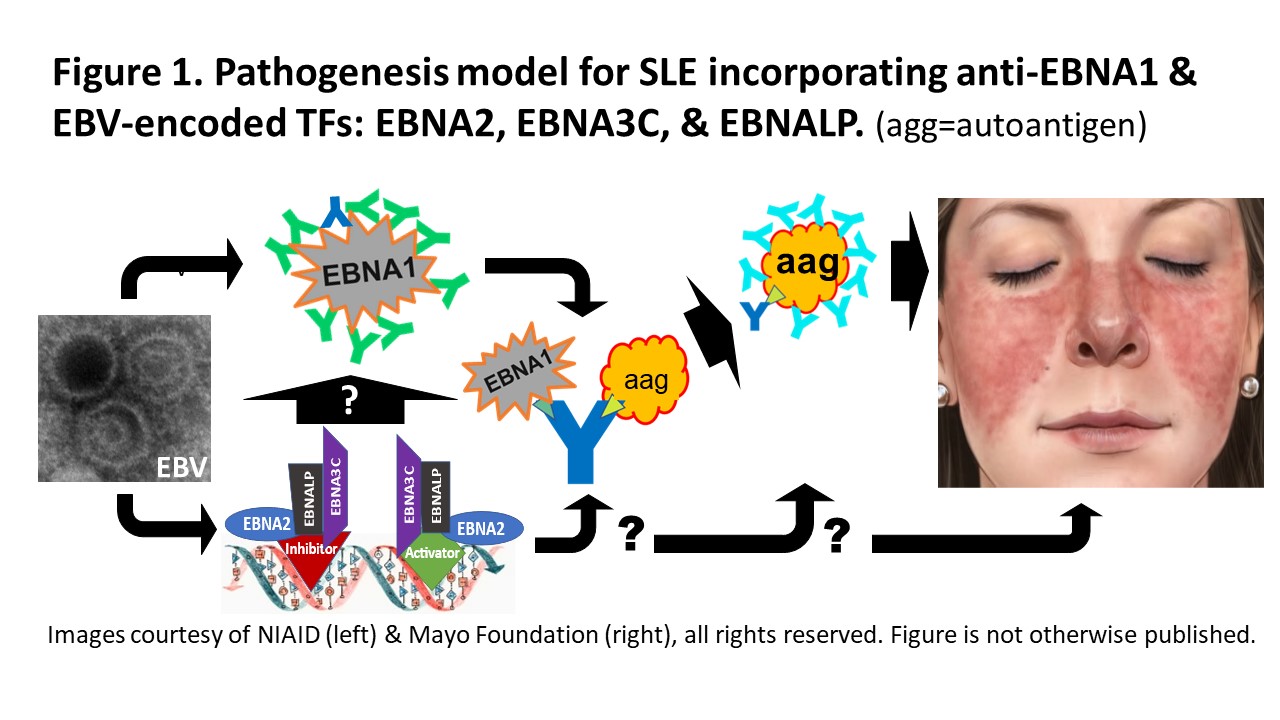
Background/Purpose: Association of EBV infection with SLE, data suggesting an anti-EBNA1 molecular mimicry fostering SLE autoimmunity, and mechanisms in EBV infected B cells support a model for EBV inducing SLE autoimmunity and clinical illness (Figure 1).
Methods: Literature review identified SLE risk loci at p< 5×10-8. We curated 9,862 ChIP-seq (Chromatin ImmunoPrecipitation with DNA sequencing) datasets from 1,339 transcription factors (TFs) in 3,531 “cellular sources” and assessed TF binding to DNA with Multimode RELI (Regulatory Element Locus Intersection (modified from PMID: 29662164)).
Results: 330 SLE risk loci (p< 5×10-8) were published prior to 2023. Of these, 255 are found in East Asians (EAS), 106 in Europeans (EU), 11 in African-Americans, and 18 in mixed Americans. 77 of 330 have been established at p< 5×10-8 in ≥3 separate studies. 40 of the 330 loci are established independently in both EAS & EU ancestries. KEGG analysis of candidate target genes and genes located near the top variant of the 330 loci, show strong associations related to, among others, IFNγ, IL-12, IL-23, B-cell receptor, & EBV pathways (6.1< OR< 16, 10-30< pa< 10-8).
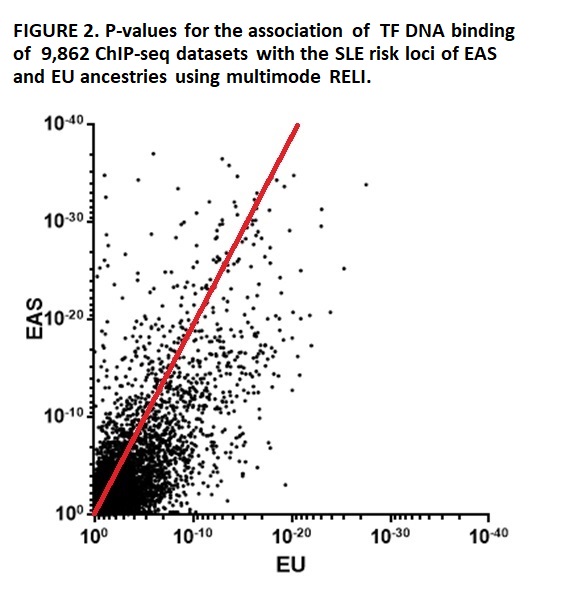
Enrichment of TFs binding to SLE loci, with disequilibrium to r2>0.8, using ChIP-seq datasets separately in EAS & EU ancestries, reveals that 200 (14.9%) of 1,339 tested TFs found in 1,871 (19.0%) of the 9,862 TF ChIP-seq datasets are associated with SLE loci at pc< 10-6 in both ancestries. The TF dataset association ranks are highly similar in EAS & EU ancestries (r=0.73 (Spearman), p< 0.0001) (Figure 2).
Of the 3,531 cellular sources, EBV-infected and transformed human B cells are most closely associated with both the EAS & EU risk loci (OR >19, p< 10-300), compared to all other cellular sources, even including B cells that are not EBV-infected. The proportions of significant (pa< 10-6) TF ChIP-seq datasets in all other tested infections are negatively associated in aggregate (OR< 0.17, p< 5.3×10-34), as are cellular sources with no known infection (OR< 0.36, p< 3×10-112).
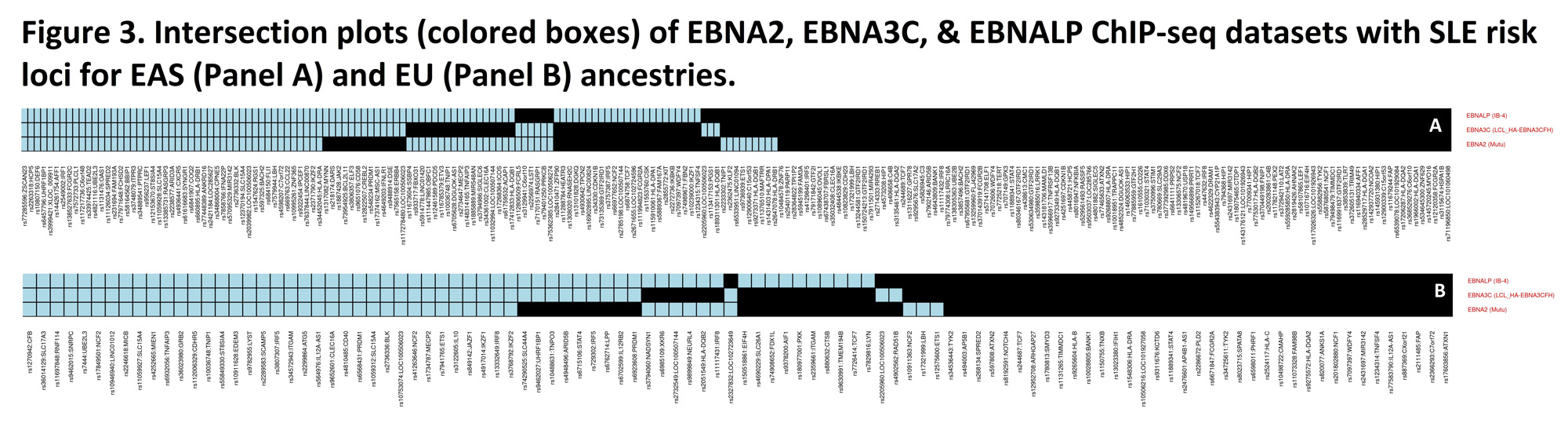
Of the associated host DNA binding of TFs at pa< 10-6, EBNA2, EBNALP, & EBNA3C, all EBV-encoded Latency III expression products, are separately associated (2.1< OR< 2.6, p< 10-11) with the SLE risk loci in both EAS & EU ancestries (Figure 3). At OR >3.2, p≤10-4, human TFs, known to form super-enhancers in EBV-infected, Latency III expressing transformed B cells, are enriched among the significant TF ChIP-seq datasets, compared to uninfected B cells independently at the EAS & EU loci. When the SLE risk loci bound by all 3 of the most closely associated EBV-encoded TF datasets in EBV transformed B cells are removed (see Figure 3), then >95% of formerly significant (pa< 10-6) TF ChIP-seq data sets are no longer significant in either the EAS or EU ancestry, which would also be true for many of the significant human-encoded TF datasets.
Conclusion: The genetic mechanisms of SLE appear similarly dependent upon EBV-encoded TF gene regulation operating in EBV-transformed B cells in both EAS and EU ancestries. The distortion of gene expression, possibly driven through EBV-encoded TFs, is present in both EAS and EU ancestries and potentially important in the pathogenesis of SLE.
To cite this abstract in AMA style:
Laurynenka V, Chen X, Parameswaran s, Kottyan L, Weirauch M, Chepelev I, Kaufman K, Harley J. The 330 Genetic Risk Loci of Systemic Lupus Erythematosus (SLE) Now Known Are Consonant with Multiple Causal Mechanisms Involving Epstein-Barr Virus (EBV)-Encoded Transcription Co-factors (TFs) in EBV-Infected B Cells [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/the-330-genetic-risk-loci-of-systemic-lupus-erythematosus-sle-now-known-are-consonant-with-multiple-causal-mechanisms-involving-epstein-barr-virus-ebv-encoded-transcription-co-factors-tfs-in-ebv/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/the-330-genetic-risk-loci-of-systemic-lupus-erythematosus-sle-now-known-are-consonant-with-multiple-causal-mechanisms-involving-epstein-barr-virus-ebv-encoded-transcription-co-factors-tfs-in-ebv/