Session Information
Date: Sunday, November 17, 2024
Title: Systemic Sclerosis & Related Disorders – Basic Science Poster I
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: Systemic sclerosis (SSc) is a rare multisystem autoimmune disease that disproportionately affects African Americans (AA). Previous work from our lab has suggested a pivotal role of HLA-II genes in SSc pathogenesis. These HLA alleles encode variations in the antigen-binding grooves of the HLA molecules that determine their binding affinity for specific antigens. Antigenic peptides presented by the HLA molecules are recognized by T cell receptors (TCRs) leading to an antigen-specific response. We hypothesized that for a rare disease like SSc, where HLA genes play an important role, there must exist a unique set of TCRs that lead to SSc pathogenesis. In this study, we examined the TCRb sequences of AA subjects with SSc and identified groups of patients who share a TCR motif that defines a unique clone in each patient.
Methods: TCRb region was deep sequenced for 132 AA subjects with SSc and 49 AA control subjects using the immunoSEQ assay (Adaptive Biotechnologies, USA). The SSc subjects were enrolled at the 24 academic centers participating in the Genome Research in African American Scleroderma Patients (GRASP) consortium. Data were analyzed using Immunarch software for repertoire analysis. TCRb clones were identified as “unique” if they were found in only one individual. GLIPH2 (Grouping of Lymphocyte Interactions by Paratope Hotspots) software was used to identify motifs that cluster the TCRs into shared antigen specificity groups.
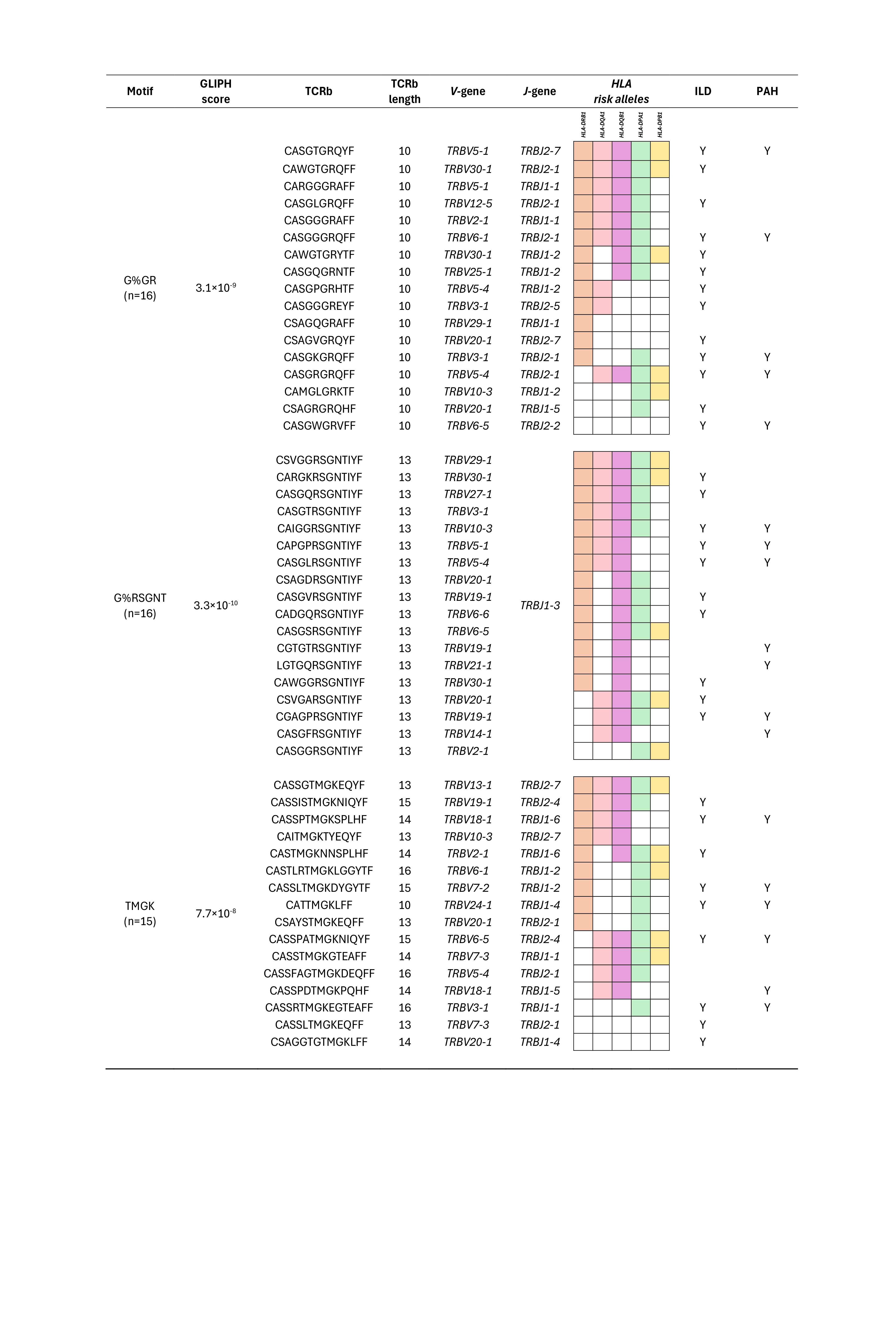
Results: We identified a total of 3,321,168 TCRb clones and using GLIPH2 (Figure 1) prioritized 228 motifs found exclusively in SSc patients and not in control subjects with significant GLIPH2 scores. Next, we identified motifs that comprised at least 10% of the samples and narrowed down to three motifs. These three motifs included TCRb clones seen in 47 SSc patients (36%) out of a total of 132 patients (Figure 2). We observed that greater than 90% of SSc patients carrying these unique clones also carried the previously reported HLA risk alleles in SSc. These unique TCRb clones were of shorter lengths (10-16 amino acids), as observed in our overall cohort analysis. There was also a preferred usage of the amino acid Glycine at position 114 in the “G%GR” motif and patients with this motif had a higher prevalence (75%) of interstitial lung disease (ILD), compared to 63% in overall SSc cohort. Motif “G%RSGNT” had Asparagine at position 114 and patients with this motif had a higher prevalence (38%) of pulmonary arterial hypertension (PAH), compared to 28% in overall SSc cohort.
Conclusion: We have identified unique TCR clones forming a motif in groups of SSc patients. The presence of these TCRb clones and skewing in the phenotype of these patients suggests a pathogenic role for these clones. Further studies are needed to understand the role of these clones in loss of tolerance.
Y indicates Yes; Blank indicates No or missing information.
To cite this abstract in AMA style:
Kaundal U, Borden C, Teehan D, Dulek B, Lack J, Shah A, Mayes M, Shriner D, Doumatey A, Bentley A, Domsic R, Medsger, Jr T, Ramos P, Silver R, Steen V, Varga J, Hsu V, Saketkoo L, Khanna D, Schiopu E, Gordon J, Criswell L, Gladue H, Derk C, Bernstein E, Bridges S, Shanmugam V, Chung L, Kafaja S, Jan R, Trojanowski M, Goldberg A, Korman B, Naz F, Dell'Orso S, Adeyemo A, Remmers E, Rotimi C, Wigley F, Boin F, Kastner D, Gourh P. TCR Motifs Identify Unique Clones in African Americans with Systemic Sclerosis [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/tcr-motifs-identify-unique-clones-in-african-americans-with-systemic-sclerosis/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/tcr-motifs-identify-unique-clones-in-african-americans-with-systemic-sclerosis/