Session Information
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: Rheumatoid arthritis (RA) is a systemic autoimmune disease characterized by a variable response to therapies, reflecting its complex and incompletely understood pathogenesis. Emerging research suggests a more prominent role for neutrophils in the inflammatory process of RA. Here, we examine alternative splicing, which can alter protein function and/or localization, to investigate the role of neutrophils in RA.
Methods: Neutrophils were isolated from 15 participants with seropositive RA. RNA was extracted, sequenced, and aligned to the reference genome (GRCh38.p14). We performed differential gene analysis of RA versus healthy neutrophils. Next, we used SplAdder to quantify the 6 types of alternative splicing events: exon skip, alternative 3’ splice site, alternative 5’ splice site, intron retention, mutually exclusive exons, and multiple exon skip. We then applied an in-silico pipeline, RiboSplitter, that we developed to predict protein changes from splicing events. The pipeline performs statistical testing, filtering, protein change prediction, and visualization. We used a beta binomial model for hypothesis testing (alpha=0.05), and p values were adjusted for multiple comparisons. The percent spliced-in (PSI) was calculated by the number of reads supporting one isoform out of the sum of both isoforms. Statistically significant events were filtered for events with greater differences between the groups. Finally, we visualized protein changes between the splicing events.
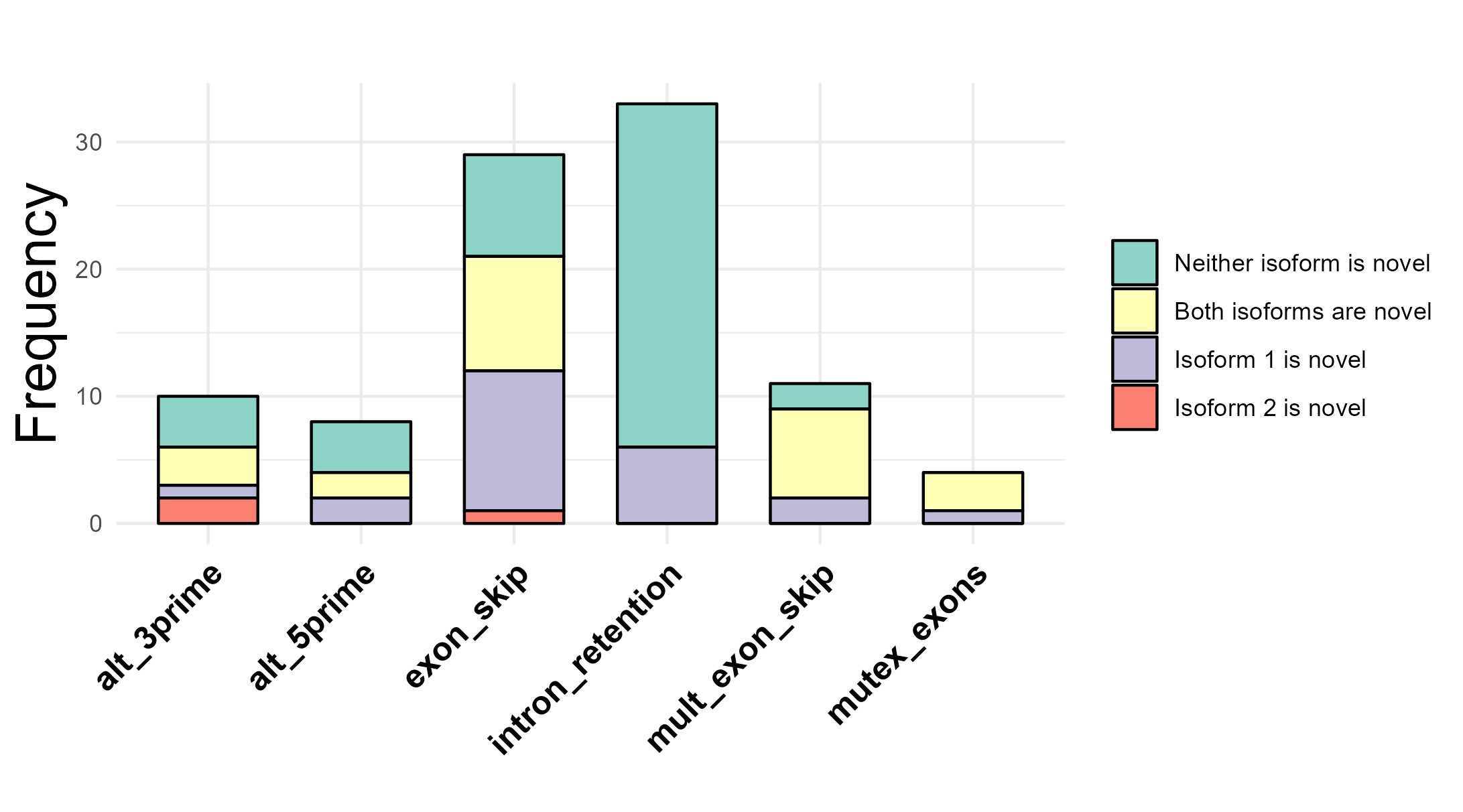
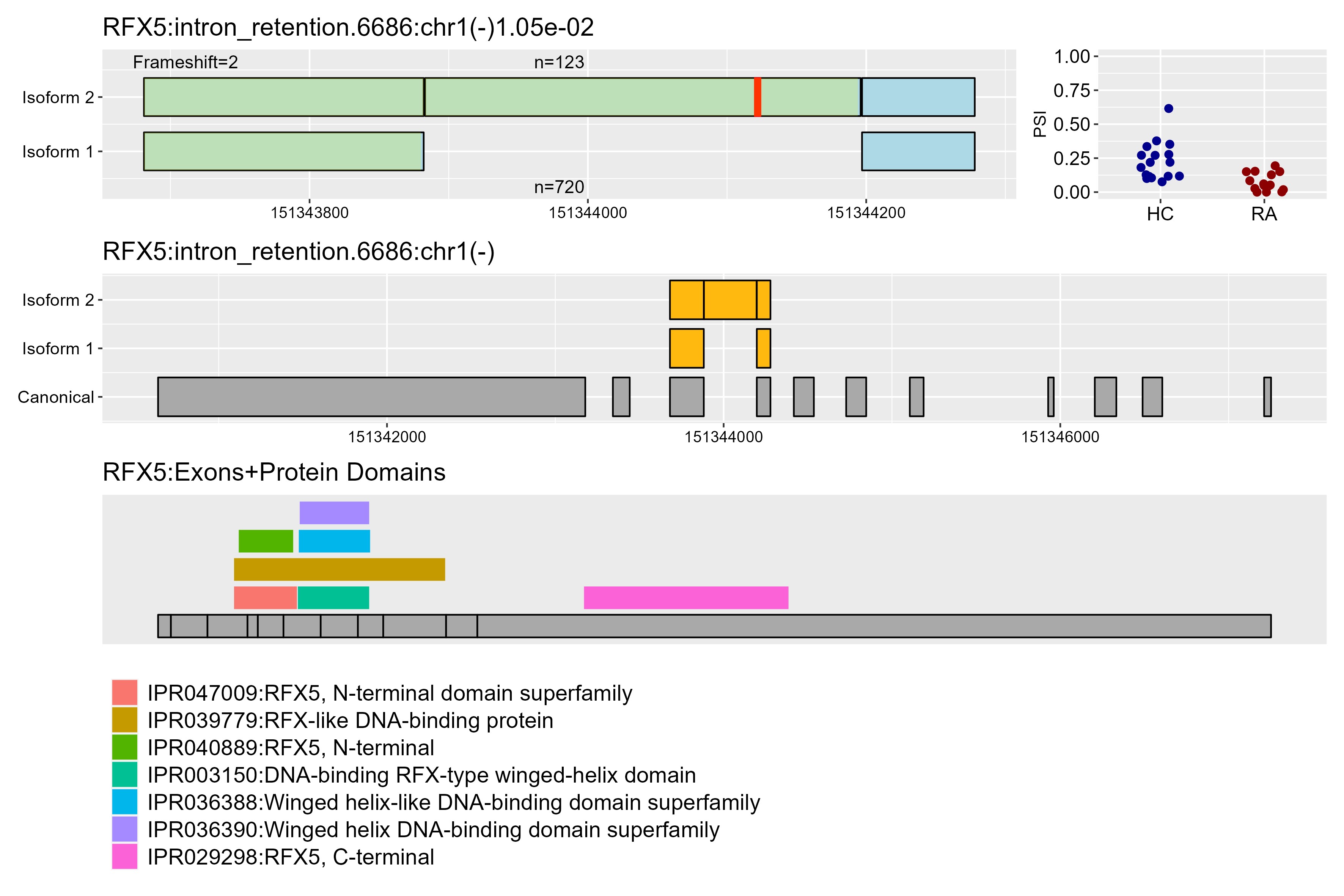
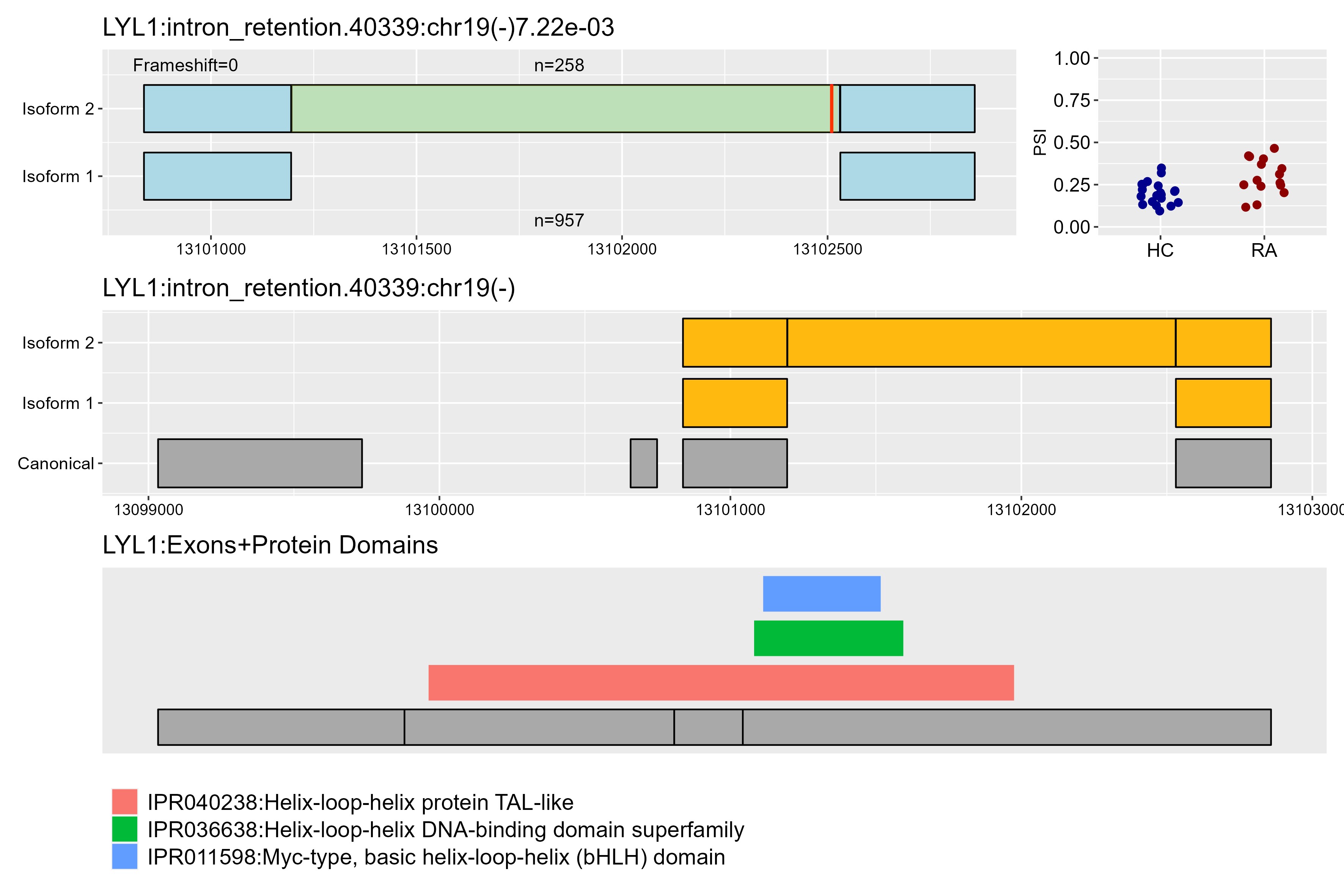
Results: While there were only three differentially expressed genes in RA compared to healthy neutrophils, we found 459 statistically significant alternative splicing events. After further filtering, we determined 95 splicing events in 83 unique genes. Intron retention was the most frequent type of event, and more than half had at least one novel isoform (Fig 1). Of particular interest, we highlighted two genes, regulatory factor X5 (RFX5) and LYL1 basic helix-loop-helix family member (LYL1). In RFX5, which is involved in the regulation of MHCII, RA samples had a higher proportion of the complete isoform, while the HC samples had more of the isoform with a stop codon, likely disrupting the DNA binding domain and C terminal domain (Fig 2). In LYL1, a transcription factor highly expressed in hematopoietic cells, RA samples had a higher proportion of isoform 2, introducing a stop codon in the retained intron shortly after the 1st exon, likely resulting in a truncated isoform that lacks the functional domains encoded by later exons (Fig 3).
Conclusion: RA neutrophils exhibited many changes in alternative splicing, but only showed few alterations in overall gene expression, emphasizing the significance of studying RNA biology beyond gene expression. We discovered multiple alternative splicing events in RA neutrophils with potential mechanistic significance. These events included alternative splicing in genes previously linked to RA, such as RFX5, which was shown to be involved in a metabolic adaptation in RA macrophages. These findings suggest alternative splicing as a promising mechanism in RA pathogenesis, highlighting the need for further research into its functional consequences and potential therapeutic targets.
Middle row: Zoomed-out view of the alternative splicing event fit to the canonical transcript.
Bottom row: Exons aligned to protein domains of the gene.
Middle row: Zoomed-out view of the alternative splicing event fit to the canonical transcript.
Bottom row: Exons aligned to protein domains of the gene.
To cite this abstract in AMA style:
Najjar R, Rogel N, Mustelin T. Splicing into Action:Investigating the Role of Alternative Splicing in Rheumatoid Arthritis Neutrophils [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/splicing-into-actioninvestigating-the-role-of-alternative-splicing-in-rheumatoid-arthritis-neutrophils/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/splicing-into-actioninvestigating-the-role-of-alternative-splicing-in-rheumatoid-arthritis-neutrophils/