Session Information
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: TLR8 has been implicated in adverse outcomes of human pregnancy. We previously reported a mouse model of spontaneous aPL-induced pregnancy loss in Sle1 mice expressing a human TLR8 transgene (Sle1.hTLR8tg). These mice experience dystocia, fetal resorptions, and placental developmental abnormalities, with both the Sle1 and hTLR8 loci required for the phenotype. Growth defects occur as early as embryonic day (e)8.5; bulk RNA-seq and flow cytometry revealed immune dysregulation, including decreased expression of IL15, decreased uterine (u)NK cells and increased myeloid cells as well as junctional zone thinning and placental ischemia. It is unclear how hTLR8 initiates the signaling cascade that leads to these changes in immune dynamics within Sle1.hTLR8tg placentas, or how other placental cell populations interact with uterine NK cells to regulate their recruitment and function.
Methods: To better define the spatial relationships during Sle1.hTLR8tg pregnancy, placental tissue was harvested from Sle1 and Sle1.hTLR8tg mice at e9.5 (decidualization and spiral artery remodeling occurring) and e13.5 (fully formed placenta). Spatial transcriptomics (n=2-3 per group, per timepoint) was performed using the 10x Xenium platform and a predesigned Mouse Tissue Atlas panel (379 gene targets) together with a custom panel (89 immune genes, including hTLR8).
Results: K-Means clustering identified major placental regions, including decidua, junctional zone, labyrinth, and mesometrial lymphoid aggregate of pregnancy (MLAp) (Fig.1A). hTLR8 was expressed only in Sle1.hTLR8tg placentas, primarily in decidua and MLAp, and in low amounts in the junctional zone and labyrinth (Fig.1B). hTLR8 expression co-localized preferentially with Itgam (CD11b) (Fig.2A) but also with a small number of stromal cells. At e9.5, Sle1.hTLR8tg placentas displayed clusters of immune cells (Cd8a, Ifng, Ncr1, Klrd1, Klrb1c, Itgam) at the junction of the MLAp and decidua (Fig.2B) not observed in Sle1 placentas and persisting through d13.5, suggesting an inflammatory/cytotoxic phenotype. An increase in placental CD8 cells was confirmed by flow cytometry. NK cell receptors (Klrb1c, Ncr1, Klrd1) and granzymes showed variable downregulation patterns, depending on the sub-type and placental region (Fig.3A-B) Ifng and Cd8a were expressed both in the MLAp and decidua (Fig.3C).
Conclusion: Several unique uNK cell subsets populate mouse and human placentas. The different distribution and variable downregulation of NK cell receptors and granzymes in our spatial data suggests a greater degree of uNK cell heterogeneity than previously appreciated. Future studies include understanding if these NK receptor expression patterns reflect different functions (e.g. differential granzyme production) and looking for potential correlations with human uNK subsets.Additionally, increased expression of Cd8a and Ifng in the MLAp and their close proximity to clusters of myeloid and uNK cells suggests a pro-inflammatory environment by which myeloid cells are activated via hTLR8 and activate CD8 cells to kill target cells such as stromal and uNK cells that are required for production of key cytokines and placental growth factors.
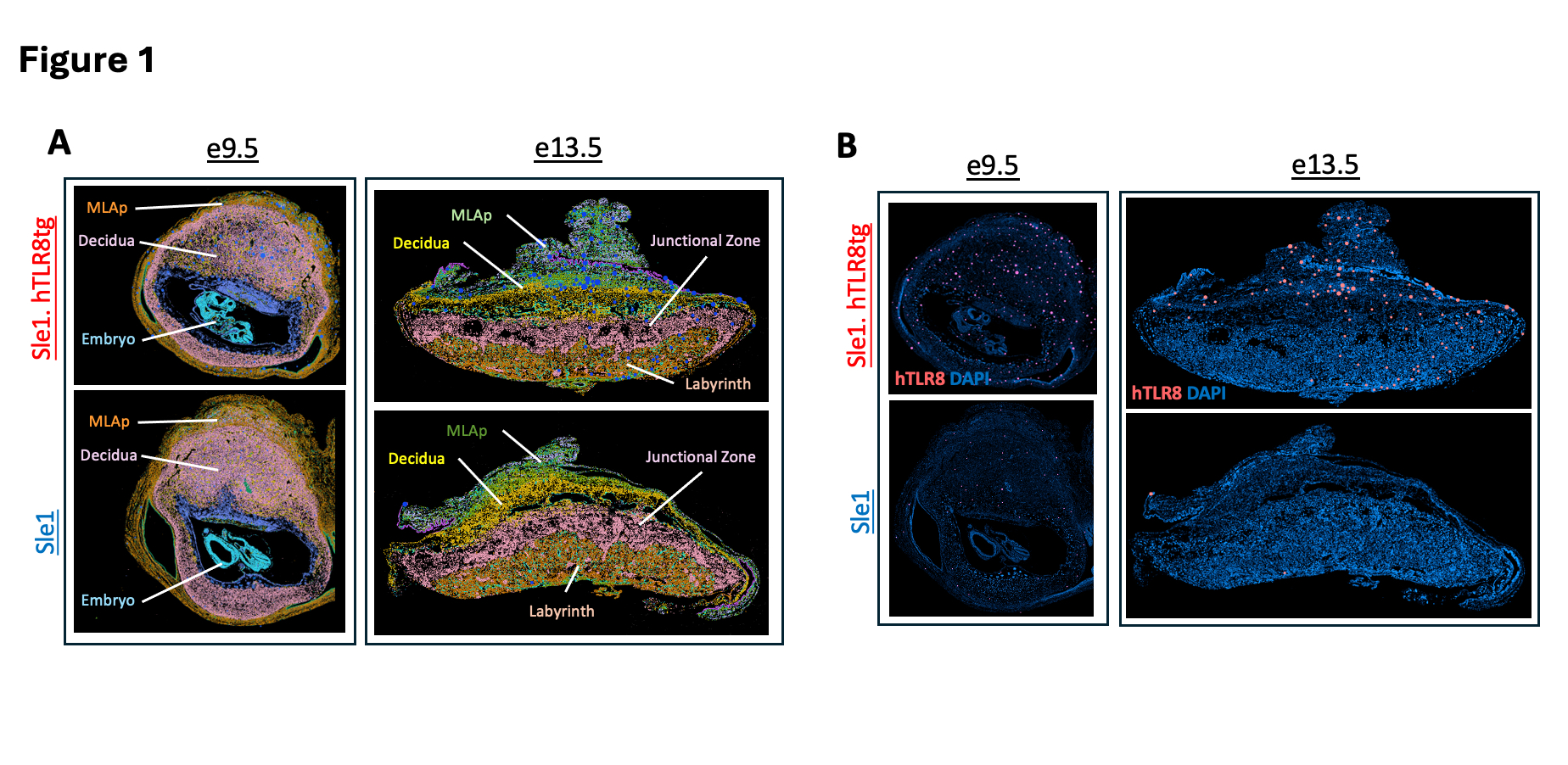 Figure 1. Spatial transcriptomics of e9.5 and e13.5 placentas in Sle1 and Sle1.hTLR8tg mice. A. K-Means Clustering at e9.5 (k=7) and at e13.5 (k=8) corresponds with distinct placental layers and cells types present during each stage of placental development. B. hTLR8 expression (red) with DAPI staining (blue) in Sle.1hTLR8 and Sle1 placentas at e9.5 (left) and e13.5 (right).
Figure 1. Spatial transcriptomics of e9.5 and e13.5 placentas in Sle1 and Sle1.hTLR8tg mice. A. K-Means Clustering at e9.5 (k=7) and at e13.5 (k=8) corresponds with distinct placental layers and cells types present during each stage of placental development. B. hTLR8 expression (red) with DAPI staining (blue) in Sle.1hTLR8 and Sle1 placentas at e9.5 (left) and e13.5 (right).
.jpg) Figure 2. Co-localization of hTLR8 and immune-related genes suggests inflammatory milieu in Sle1.huTLR8tg placentas. A. Sle1.huTLR8tg placentas at e9.5 (top) and e13.5 (bottom) with inlay showing co-expression of hTLR8 (pink) with Itgam (gold-CD11b). B. Local clusters of NK cells (represented by granzyme and NK cell receptor genes), myeloid cells (represented by Itgam and Itgax), along with Ifng, Cd8a, and hTLR8 in e9.5 Sle1.huTLR8tg placenta.
Figure 2. Co-localization of hTLR8 and immune-related genes suggests inflammatory milieu in Sle1.huTLR8tg placentas. A. Sle1.huTLR8tg placentas at e9.5 (top) and e13.5 (bottom) with inlay showing co-expression of hTLR8 (pink) with Itgam (gold-CD11b). B. Local clusters of NK cells (represented by granzyme and NK cell receptor genes), myeloid cells (represented by Itgam and Itgax), along with Ifng, Cd8a, and hTLR8 in e9.5 Sle1.huTLR8tg placenta.
.jpg) Figure 3. Immune cell-related gene expression patterns in Sle1 and Sle.1hTLR8tg placentas at e9.5. A. Granzyme genes were all downregulated in Sle1.hTLR8tg placentas compared with Sle1 placentas, but Gzmb downregulation was markedly different between the MLAp (where it was virtually absent) and the decidua (with only a modest decrease in expression). B. NK cell receptor genes were similarly downregulated in Sle1.hTLR8tg placentas, but Klrb1c showed the largest decrease whereas Klrd1 remained unchanged. Expression of Ncr1 and Klrb1c differed between the MLAp and decidua regions, suggesting a distinct composition of uNK subsets in each region. C. Ifng and Cd8a expression were both increased in Sle1.hTLR8tg placentas, particularly in the MLAp, with the large majority of Ifng co-localizing with Cd8a+/Itgam-/Itgax- cells (as opposed to cells expressing NK cell receptor genes), showing that CD8+ T cells are the main producers of IFNg at e9.5 in Sle1.hTLR8tg placentas.
Figure 3. Immune cell-related gene expression patterns in Sle1 and Sle.1hTLR8tg placentas at e9.5. A. Granzyme genes were all downregulated in Sle1.hTLR8tg placentas compared with Sle1 placentas, but Gzmb downregulation was markedly different between the MLAp (where it was virtually absent) and the decidua (with only a modest decrease in expression). B. NK cell receptor genes were similarly downregulated in Sle1.hTLR8tg placentas, but Klrb1c showed the largest decrease whereas Klrd1 remained unchanged. Expression of Ncr1 and Klrb1c differed between the MLAp and decidua regions, suggesting a distinct composition of uNK subsets in each region. C. Ifng and Cd8a expression were both increased in Sle1.hTLR8tg placentas, particularly in the MLAp, with the large majority of Ifng co-localizing with Cd8a+/Itgam-/Itgax- cells (as opposed to cells expressing NK cell receptor genes), showing that CD8+ T cells are the main producers of IFNg at e9.5 in Sle1.hTLR8tg placentas.
To cite this abstract in AMA style:
Xia Y, Hoover P, Arazi A, Davidson A. Spatial Transcriptomics Reveal Altered Immune Dynamics Regulating Placental Development In a Humanized-TLR8 Mouse Model of Spontaneous Anti-Phospholipid Antibody Induced Pregnancy Loss [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/spatial-transcriptomics-reveal-altered-immune-dynamics-regulating-placental-development-in-a-humanized-tlr8-mouse-model-of-spontaneous-anti-phospholipid-antibody-induced-pregnancy-loss/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/spatial-transcriptomics-reveal-altered-immune-dynamics-regulating-placental-development-in-a-humanized-tlr8-mouse-model-of-spontaneous-anti-phospholipid-antibody-induced-pregnancy-loss/
