Session Information
Session Type: Abstract Session
Session Time: 4:00PM-5:30PM
Background/Purpose: Recent single-cell RNA sequencing (scRNA seq) data has been generated across several tissues in autoimmune diseases, helping to understand the cellular and molecular underpinnings of disease. However, the location of cells or interest identified and their proximity to important structure in the tissues, has not been as well described. Pairing spatial transcriptomics (ST) with scRNA seq data would add the much needed in situ spatial context of cellular and extracellular matrix tissue deposition, provide detail to characterize ligand-receptor and cell-cell interaction patterns and has the potential for biomarker discovery for rare diseases like scleroderma. Our scleroderma cohort, both localized scleroderma (LS) and systemic sclerosis (SSc) patients, have two 4mm skin punch biopsies collected in tandem from same site, one sample processed with scRNAseq and the other formalin-fixed paraffin-embedded (FFPE) available for spatial transcriptomics application.
Methods: To preserve both the cellular location and architecture of the tissue while also gathering a large quantity of information about the cellular phenotypes of cells, we applied an innovative technology, VisiumSpatialGeneExpression(10XGenomics), Visium CytAssist for formalin-fixed paraffin-embedded (FFPE), which captures transcripts on spatially barcoded slide with a probe panel targeting 18,085 human protein coding genes. After sequencing, barcode information was used to determine the transcriptional profile (cellular representation) of each spot within the spatial context of the entire tissue(SpaceRanger, 10x Genomics). A novel integrative analyses approach was then applied that combines scRNA seq with spatial location of cells with in the tissue, Robust Cell-type Decomposition, which is an optimized deconvolution pipeline to decipher specific cell-type composition by anatomic location and can obtain details to the individual cell level.
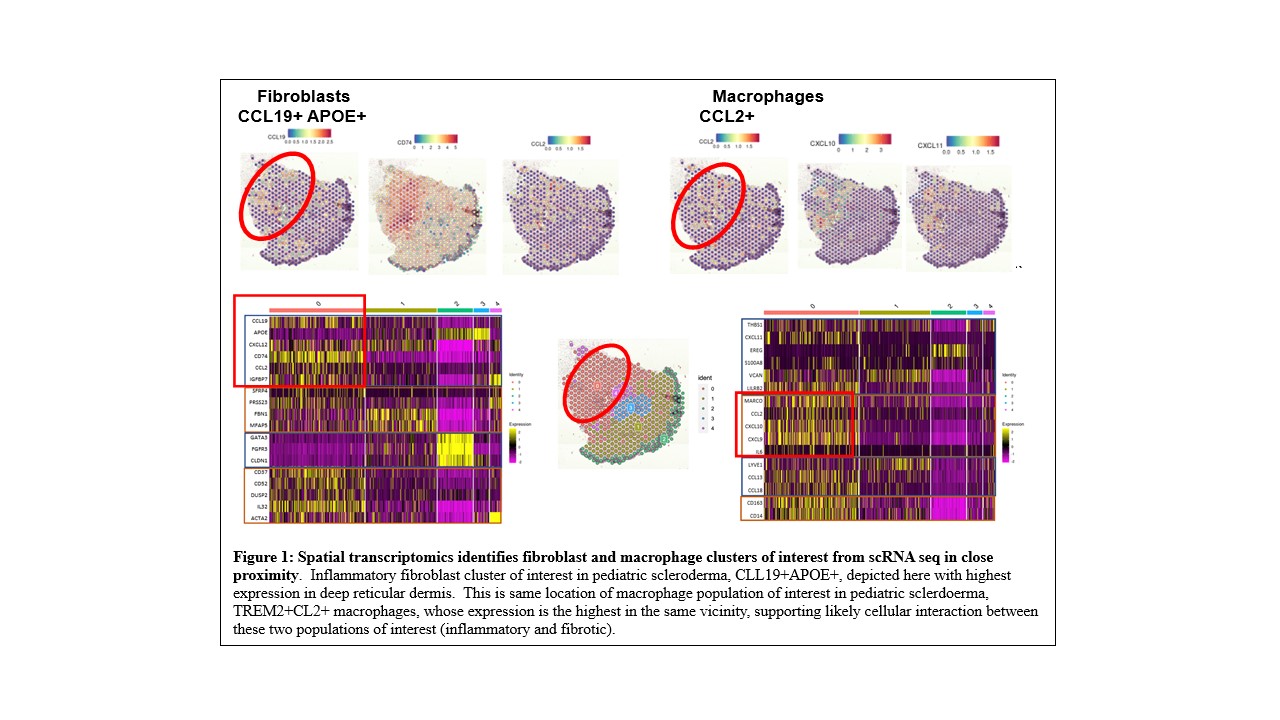
Results: We performed a pilot study on 4 skin FFPE samples, 3 from pediatric LS and 1 from pediatric SSc patients using the Visium 10X CytAssist for FFPE platform. We obtained an average of a total number of 566 spots under tissue section (4 mm x 4mm) with mean reads per spot 257,119, with 96% reads mapped to probe set, with 2,896 median genes detected per spot, and a total number of 18,029 genes detected. Transcriptomic expression using either marker genes or unbiased clustering was able to distinguish epidermis, dermis, and inflammatory regions. We also investigated some of the cell subsets and upregulated genes in those clusters from our scRNA seq data and observed the co-localization of an inflammatory fibroblast (CL19+APOE+) and a macrophage population (CCL2+TREM2+) that highly expresses IL-β in pediatric scleroderma together in the deep reticular dermis (Figure 1).
Conclusion: Using an innovative bioinformatic approach coupling scRNA seq and ST from paired tissue samples, we are able to identify dysregulated genes of interest, cells expressing these genes and their location within the skin and surrounding cells. To our knowledge, this would be the first study to employ spatial transcriptomics in localized scleroderma and systemic sclerosis skin.
To cite this abstract in AMA style:
Torok K, Lai Y, Xu Z, Sanyal A, Werner G, Hutchins T, Cheng C, Chen W. Spatial Transcriptional Alterations in the Cellular Landscape of Pediatric Scleroderma Skin [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/spatial-transcriptional-alterations-in-the-cellular-landscape-of-pediatric-scleroderma-skin/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/spatial-transcriptional-alterations-in-the-cellular-landscape-of-pediatric-scleroderma-skin/