Session Information
Date: Saturday, November 16, 2024
Title: Abstracts: Systemic Sclerosis & Related Disorders – Basic Science
Session Type: Abstract Session
Session Time: 1:00PM-2:30PM
Background/Purpose: Interstitial lung disease encompasses a heterogenous group of conditions broadly characterized by inflammation and progressive fibrosis of the lung[1]. There remains an ongoing search for key features that drive the heterogeneity observed across ILD.
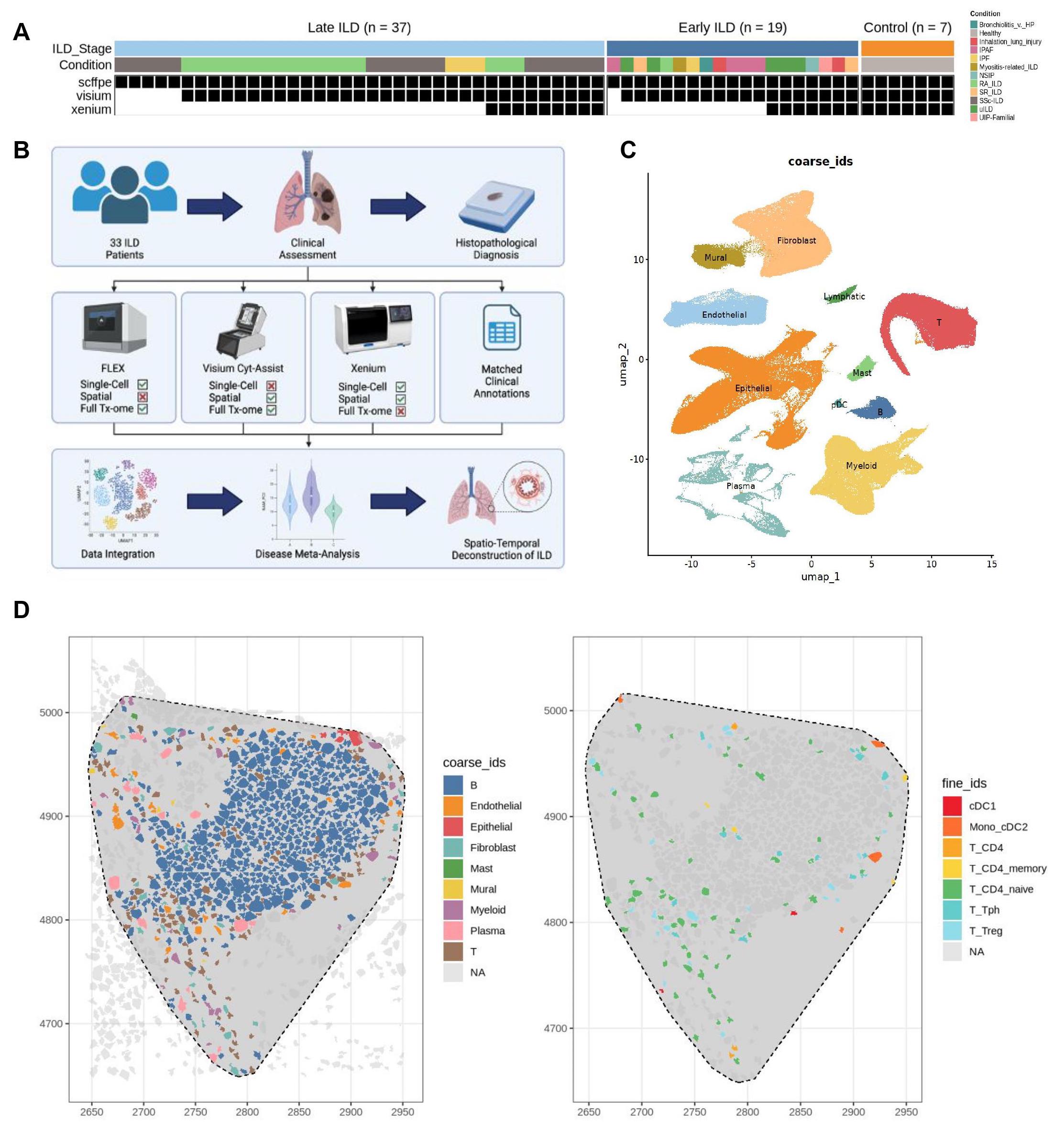
Methods: We used three complementary assays from 10x Genomics to profile single cells. The Single Cell Flex assay allows for the transcriptomic profiling of formalin-fixed paraffin-embedded (FFPE) tissue, the Visium CytAssist assay allows for spatially-resolved whole-transcriptomic profiling of tissue at a supercellular resolution, and 10x’s Xenium assay facilitates the detection of a curated panel of several hundred genes and their localizations at subcellular resolution. Anchoring our analysis on fine cell types identified in single cell Flex data, we leveraged TACCO[2], a reference-based deconvolution algorithm, and an in-house reference-based label transfer pipeline to resolve the spatial localizations of cell types in our Visium and Xenium data respectively.
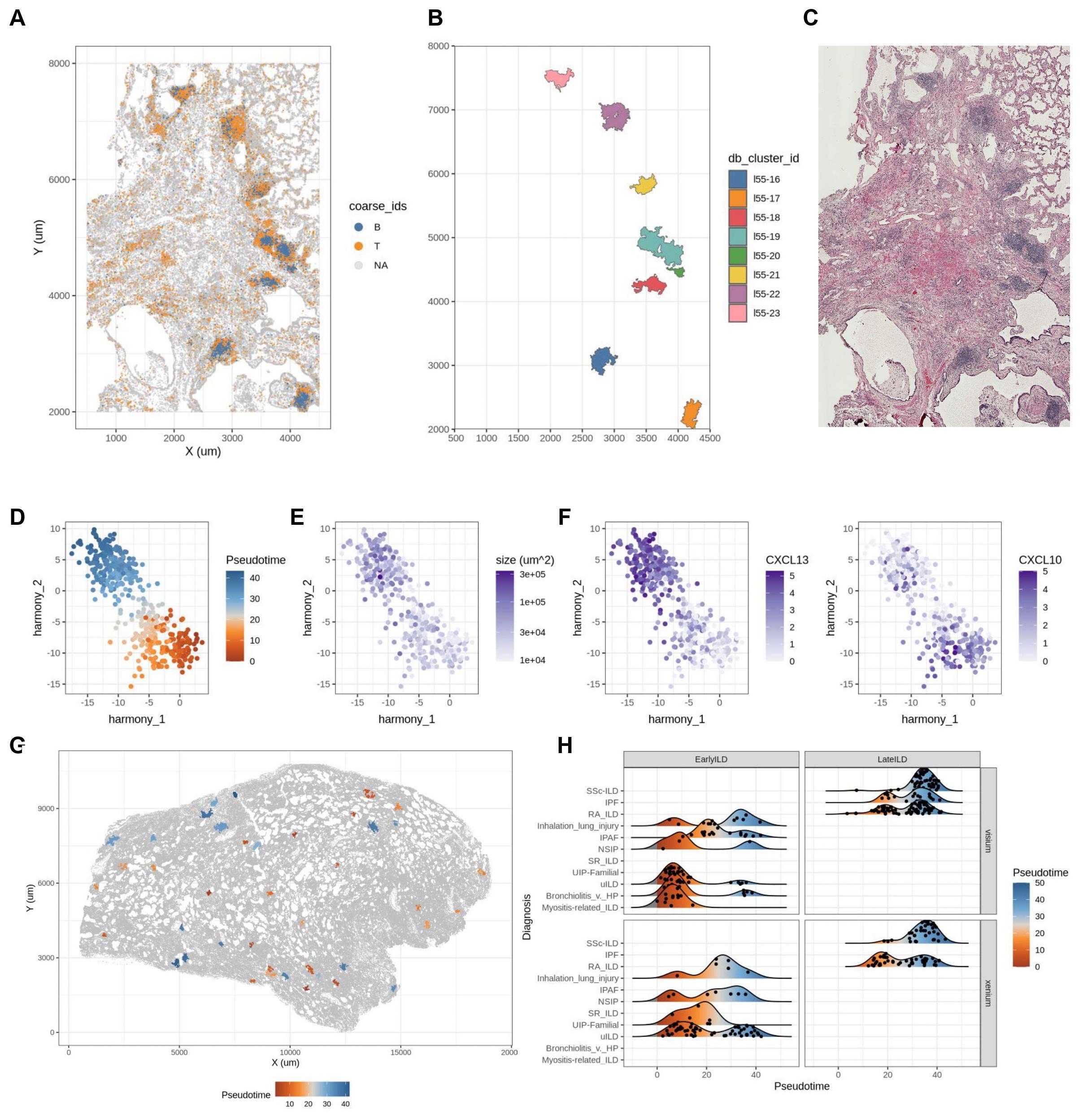
Results: With matched serial tissue sections from 63 patient samples (n = 36 rheumatic ILD, n = 20 non-rheumatic ILD, and n = 7 non-ILD controls), we generated transcriptomic data for over 12 million cells across 3 modalities (Fig 1A-C). We observed lymphocytes spatially organized into distinct immune aggregate regions across ILD samples (Fig 1D). These regions were isolated for subsequent analysis (Fig 2A-C). We used pseudotime inference analysis[3] to map individual aggregates along a transcriptional trajectory (Fig 2D), with smaller myeloid-rich aggregates on one end and larger more mature B cell-rich aggregates on the other (Fig 2E). Lymphocyte trafficking cytokines were differentially expressed across this transcriptional gradient (Fig 2F). Although most individual samples contained aggregates at multiple stages along the trajectory (Fig 2G), the mean composition of aggregates associated with clinical annotations. Patients with late-stage SSc and idiopathic pulmonary fibrosis tended to have more mature aggregates, while patients with bronchiolitis and myositis-related ILD tended to have more nascent aggregates, and those with RA-ILD and undifferentiated ILD were more heterogeneous (Fig 2H).
Conclusion: Here, we construct a single-cell and spatial atlas of ILD to illuminate the structural, cellular, and molecular landscape of the disease. Integrating these data with clinical metadata, we show: 1) pairing FFPE-compatible assays with hospital tissue archives facilitates a cross-sectional study of ILD in time and space, 2) immune aggregate regions exhibit transcriptional, compositional, and morphological patterns consistent with a complexity gradient, and 3) immune aggregate complexity is associated with clinical annotations of ILD stage and subtype.
[1] A. U. Wells, C. P. Denton, Nat. Rev. Rheumatol. 2014, 10, 728.
[2] S. Mages, N. Moriel, I. Avraham-Davidi, E. Murray, J. Watter, F. Chen, O. Rozenblatt-Rosen, J. Klughammer, A. Regev, M. Nitzan, Nat. Biotechnol. 2023, 41, 1465.
[3] K. Street, D. Risso, R. B. Fletcher, D. Das, J. Ngai, N. Yosef, E. Purdom, S. Dudoit, BMC Genomics 2018, 19, 477.
(A) Study design. (B) Experimental workflow. (B) Integrative uniform manifold and projection (UMAP) based on mRNA discriminated major cell types. (D) Spatial distribution of coarse (left) and fine (right) cell types in a representative immune aggregate region.
(A) Spatial distribution of lymphocytes in an immune aggregate rich FOV. (B) Isolated immune aggregate regions. (C) Associated H&E image of the previously shown FOV. (D) Principal component plot of pseudo bulk transcriptomic profiles of immune aggregate regions colored by predicted pseudotime ordering. (E) Immune aggregate size vs. pseudotime. (F) Normalized expression of CXCL13 (left) and CXCL10 (right) within immune aggregates. (G) Spatial distribution of immune aggregate regions in an early ILD sample colored by pseudotime. (H) Density ridge plot comparing immune aggregate pseudotime across technologies and clinical annotations.
To cite this abstract in AMA style:
Tran M, Gao C, Palshikar M, Liu J, Kamiya M, McDermott G, Joerns E, Simmons D, Gate R, Rao D, Sparks J, Kim E, Wei K, Korsunsky i. Spatial Reconstruction of Interstitial Lung Disease [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/spatial-reconstruction-of-interstitial-lung-disease/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/spatial-reconstruction-of-interstitial-lung-disease/