Session Information
Date: Monday, November 18, 2024
Title: Systemic Sclerosis & Related Disorders – Basic Science Poster II
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Long non-coding RNAs (lncRNA) are a class of transcripts that regulate gene expression. Currently, only few lncRNAs have been linked to SSc pathogenesis. As lncRNAs often evade large screens due to their high cell specificity and low copy numbers, distinct analysis methods are needed to identify differentially expressed lncRNAs. Here, we aimed to identify candidate lncRNAs that may play a role in SSc using a bioinformatic pipeline optimized for non-coding genes.
Methods: Single cell RNA sequencing (scRNA-seq) data from sex matched SSc and healthy control (HC) skin biopsies were obtained with the 10X Genomics workflow. To analyze lncRNAs expression, we optimized a scRNA-seq bioinformatic pipeline, which included STARsolo alignment and differential gene expression analysis (DGEA) with DESeq2. Human dermal fibroblasts were isolated from skin biopsies of SSc patients and HC. LncRNA expression was analyzed by RT-PCR. Cells were treated with 10 ng/mL TGF-β1. LncRNAs MIR193-BHG and LINC01503 were silenced in SSc dermal fibroblasts using locked nucleic acid antisense oligonucleotides (LNA GapmeRs), followed by bulk RNA sequencing (bulk RNA-seq) and subsequent DGEA as well as Gene Set Enrichment Analysis (GSEA).
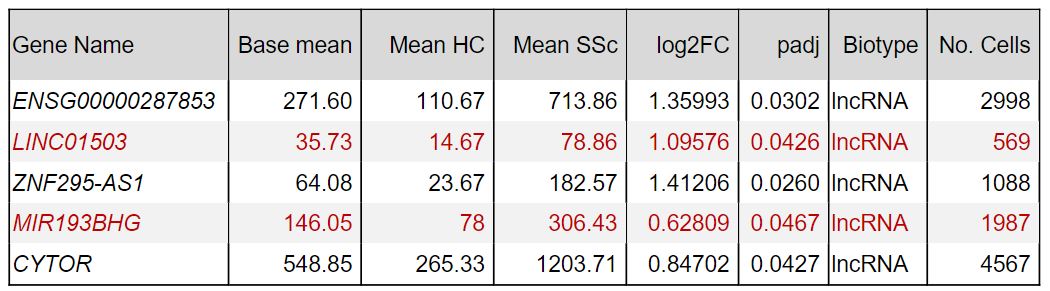
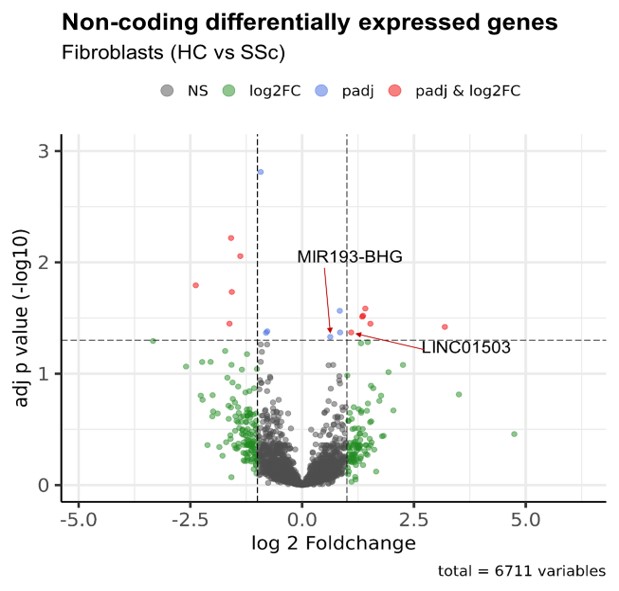
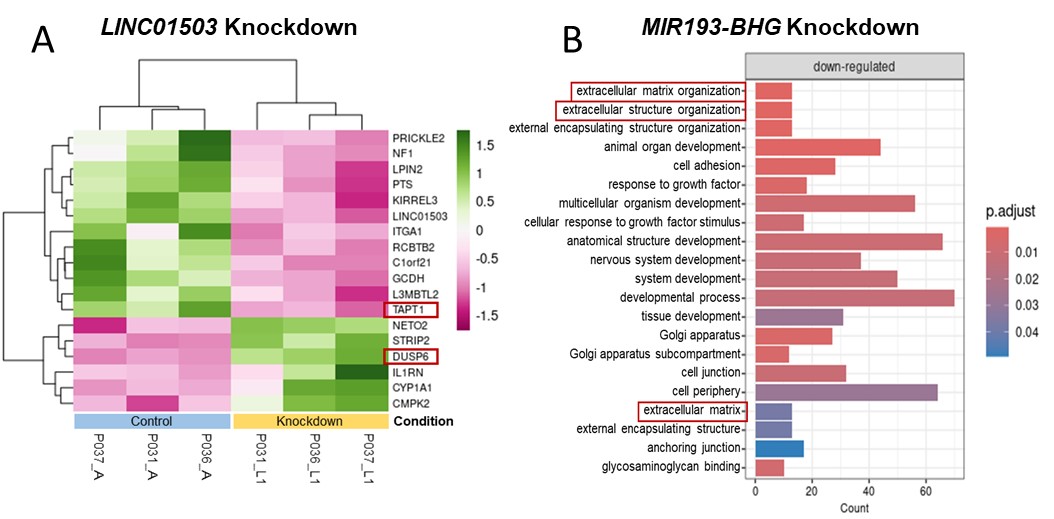
Results: Our optimized bioinformatic pipeline enabled us to account for the low expression levels and cell specificity of lncRNAs. With a focus on fibroblasts, DGEA revealed 23 differentially expressed lncRNAs from SSc patients (n=7) as compared to HCs (n=6). Pseudo bulk analysis provided the five top lncRNAs differentially expressed in SSc fibroblasts (CYTOR, ENSG00000287853, LINC01503, MIR193-BHG, ZNF295-AS1). Subsequent RT-PCR confirmation analysis in cultured fibroblasts showed a significant 1.89-fold increase of LINC01503 basal gene expression in SSc fibroblasts as compared to HC fibroblasts (n=8, p< 0.05). TGF-β1 stimulation of fibroblasts led to a 2.96-fold increase of LINC01503 gene expression in SSc cells (n=6, p< 0.01). Conversely, basal MIR193-BHG expression was not significantly altered between SSc and HC fibroblasts, MIR193-BHG expression was significantly induced by TGF-β1 stimulation 1.42-fold in HC cells (p< 0.001) and by 1.39-fold in SSc cells (p< 0.01) respectively. Knockdown of LINC01503 in SSc fibroblasts followed by bulk RNA-seq analysis showed TAPT1 and DUSP6 among the top differentially expressed genes. TAPT1 protein instability has been linked to impaired collagen fibril formation. DUSP6 negatively regulates members of the Mitogen-Activated Protein Kinase (MAPK) family. Knockdown of MIR193-BHG in SSc fibroblasts lead to the downregulation of several extracellular matrix genes (SULF1, RB1, B4GALT1, CCDC80, CRISPLD2, FBLN5, ADAMTS5, RUNX1, FLRT2, VIT). GSEA showed “extracellular matrix organization”, “extracellular structure organization” and “extracellular matrix” among the top enriched gene sets.
Conclusion: Our targeted bioinformatic analysis revealed previously overlooked lncRNAs LINC01503 and MIR193-BHG, which may play a role in SSc pathogenesis. These findings contribute to a deeper understanding of disrupted regulatory processes in SSc skin fibrosis and highlight the importance of exploring lncRNAs to better understand SSc pathomechanisms.
To cite this abstract in AMA style:
Wagner S, Pachera E, Bonazza G, Geiss C, Hofman A, Much L, Li L, Bearzi P, Hoffmann-Vold A, Distler O. Single-Cell RNA SequencingBioinformatic Pipeline Optimized for Non-Coding GenesIdentified LINC01503 and MIR193-BHG as Potential Pathogenic Effectors in Systemic Sclerosis Fibroblasts [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/single-cell-rna-sequencingbioinformatic-pipeline-optimized-for-non-coding-genesidentified-linc01503-and-mir193-bhg-as-potential-pathogenic-effectors-in-systemic-sclerosis-fibroblasts/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/single-cell-rna-sequencingbioinformatic-pipeline-optimized-for-non-coding-genesidentified-linc01503-and-mir193-bhg-as-potential-pathogenic-effectors-in-systemic-sclerosis-fibroblasts/