Session Information
Date: Sunday, November 13, 2022
Title: Systemic Sclerosis and Related Disorders – Basic Science Poster
Session Type: Poster Session C
Session Time: 1:00PM-3:00PM
Background/Purpose: Systemic Sclerosis (SSc) currently lacks reliable in vitro models of skin fibrosis constructed from all human cells. We have developed a skin-like tissue model of systemic sclerosis (SSc) that recapitulates the increased tissue thickness, stiffness, and molecular phenotype of SSc. Recent studies have identified specific pathogenic fibroblast (FB) subsets in skin and other affected tissues. The goal of this study was to characterize the cellular heterogeneity observed in our 3D tissues and compare these data to in vivo studies. We hypothesize that our 3D model will in part recapitulate the FB heterogeneity observed in human skin and provide a tool for further insight into the cell-specific mechanisms of fibrosis in SSc.
Methods: Self-assembled Skin-equivalent (saSE) 3D tissues were grown in vitro over a period of 5 weeks by seeding healthy control (HC) or SSc FBs and monocytes into transwells, feeding with autologous plasma, and then layering normal human keratinocytes (NHKs) to form an epidermis. Following collagenase digestion of tissues, multi-omic scATAC-seq and RNA-seq data were generated using 10x protocols for a total of 3 biological replicates for both HC and SSc tissues. Statistical analyses of the single-cell dataset were performed in R using the Seurat and Signac packages. Samples were integrated and the ClusTree package was used to select the appropriate clustering resolution. Cell type was determined using cell type-specific gene expression.
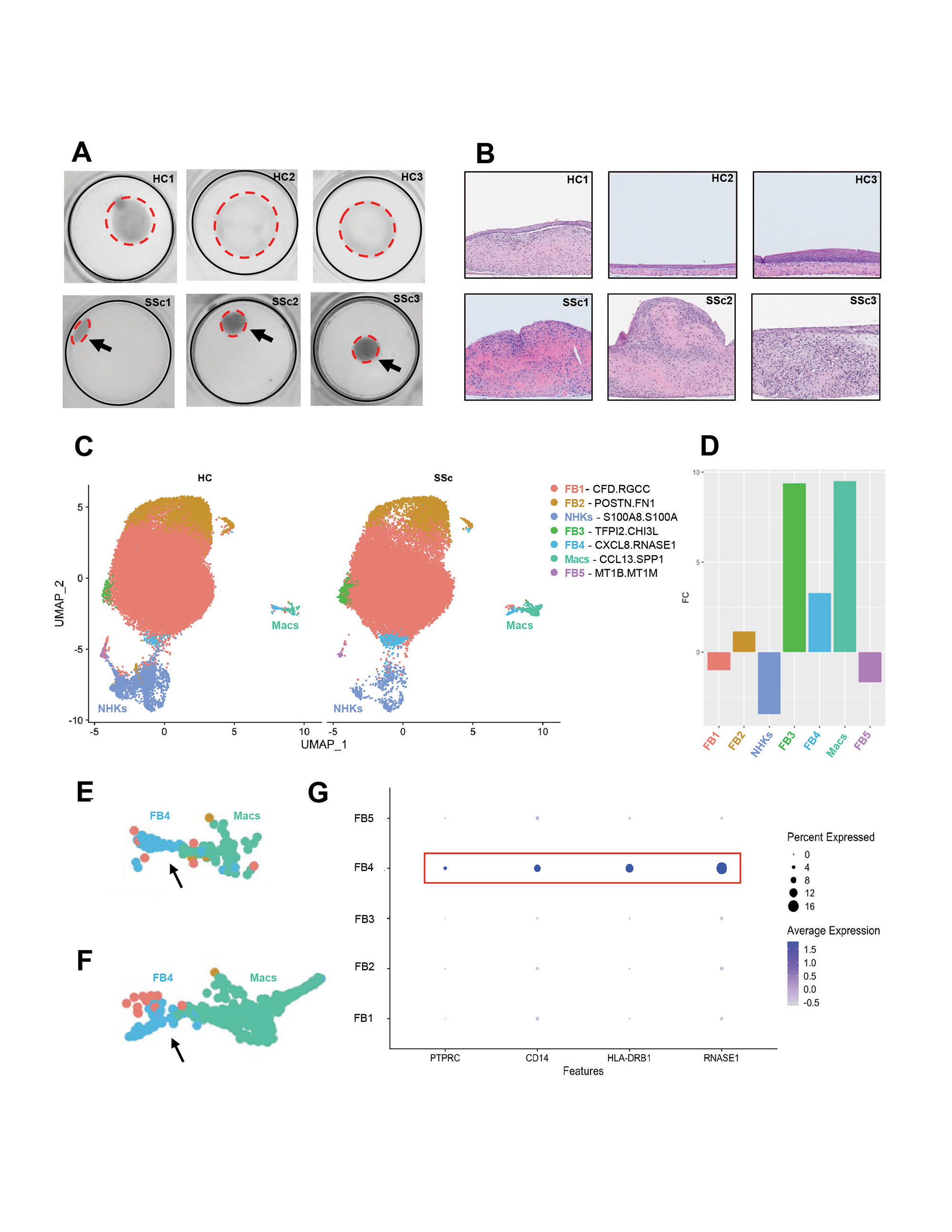
Results: Overhead images (Fig. 1A) and H&E histology (Fig. 1B) revealed a more contracted, thicker phenotype for SSc saSE skin-like tissues. Clustering resulted in identification of macrophage (Mac), NHK, and 4 major FB populations (Fig. 1C). Fold enrichment analysis showed a clear disease-specific enrichment of both Macs and 2 FB subsets (FB3 and FB5) (Fig. 1D). Cluster FB3 was the most enriched and displayed increased expression of SFRP4 and accessibility of the EGR1 and JUNB binding motifs. It was characterized by pathways associated with extracellular matrix proteins, proliferation, angiogenesis, and the recruitment of immune cells. FB5 cluster was also significantly enriched and was clustered adjacent to both FB and Mac populations (Fig. 1E-F). This population expressed collagen but was also enriched for myeloid genes including CD45, HLA-DRB1, and the monocyte marker CD14 (Fig. 1G). Pathways include wound healing, MHC II binding, and activation of T-cells. This cluster was not observed in scRNA-seq data from 3D tissues lacking the addition of monocytes.
Conclusion:
We identified 2 FB populations upregulated in the SSc saSE tissues. The FB3 subset highly expresses SFRP4 and may recapitulate a population recently identified as enriched in SSc skin via single-cell analysis. Epigenetic data suggests that EGR1 and JUNB may play a critical role in maintaining this FB state. In addition, we identify enrichment of a novel FB subset, FB5, with markers of both myeloid and mesenchymal cells. Most importantly, we were able to characterize the FB heterogeneity of our 3D skin-like tissue model confirming that this model can approximate the cellular complexity observed in human skin and may serve as a suitable model for additional studies of pathogenic FB subsets in SSc.
To cite this abstract in AMA style:
Abel T, Kosarek N, Parvizi R, Jarnagin H, Huang M, Smith A, Mariani M, Popovich D, Yang H, Wood T, Garlick J, Pioli P, Whitfield M. Single-cell Multi-omic Analysis of a 3D Skin-Like Tissue Model Provides Insights into Molecular and Cellular Drivers of Systemic Sclerosis [abstract]. Arthritis Rheumatol. 2022; 74 (suppl 9). https://acrabstracts.org/abstract/single-cell-multi-omic-analysis-of-a-3d-skin-like-tissue-model-provides-insights-into-molecular-and-cellular-drivers-of-systemic-sclerosis/. Accessed .« Back to ACR Convergence 2022
ACR Meeting Abstracts - https://acrabstracts.org/abstract/single-cell-multi-omic-analysis-of-a-3d-skin-like-tissue-model-provides-insights-into-molecular-and-cellular-drivers-of-systemic-sclerosis/