Session Information
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: We characterized the spatial distribution of immune cells and identified hub genes within activated molecular networks in key immune cell populations, based on the muscle transcriptome profile of a patient with anti-glycyl tRNA synthetase syndrome.
Methods: Single-cell and spatial transcriptome analyses were performed on muscle biopsy specimens from a male patient in his 40s diagnosed with anti-glycyl tRNA synthetase syndrome. Spatial transcriptomic profiling identified gene expression clusters corresponding to regions of intense inflammation, characterized by immune cell infiltration. Differential gene expression analysis of these immune cell populations was performed using single-cell transcriptomic data. Subsequent molecular network and topological analyses were used to identify hub genes within activated signaling pathways. Validation of hub gene expression was conducted using the publicly available dataset GSE48280, comprising transcriptomic data from patients with idiopathic inflammatory myopathies.
Results: Gene expression profiling of muscle biopsy specimens identified six spatial clusters, one of which localized to a region of pronounced inflammation. This inflammatory cluster was predominantly composed of CD14⁺CD163⁺ M2 macrophages (Figure 1). Single-cell transcriptomic analysis further delineated 10 distinct cell clusters, including one corresponding to CD163-positive M2 macrophages. Molecular network construction and topological analysis of the top 30 differentially expressed genes in the CD163-positive M2 macrophage cluster identified two hub genes (Figure 2). Validation using the GSE48280 dataset, which includes RNA microarray data from muscle biopsies of five healthy controls and five patients with dermatomyositis, demonstrated that all identified hub genes exhibited a trend toward elevated expression in dermatomyositis patients (Figure 3).
Conclusion: In myositis associated with anti-glycyl tRNA synthetase syndrome, CD163-positive M2 macrophages, characterized by a limited set of hub genes, play a pivotal role in orchestrating the local inflammatory response within affected muscle tissue.
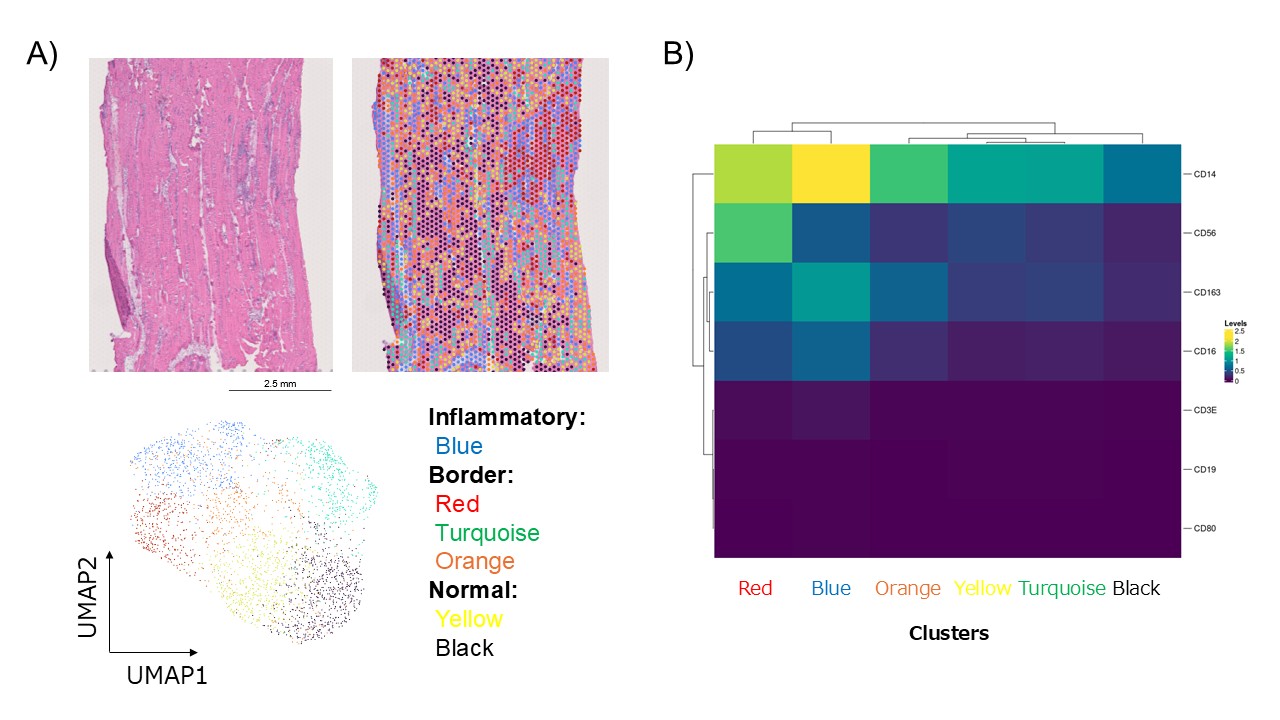 Figure 1. Spatial transcriptomic profiling. A) Six spatial clusters were identified. Blue cluster localized to a region of pronounced inflammation. B) Blue cluster was predominantly composed of CD14⁺CD163⁺ M2 macrophages.
Figure 1. Spatial transcriptomic profiling. A) Six spatial clusters were identified. Blue cluster localized to a region of pronounced inflammation. B) Blue cluster was predominantly composed of CD14⁺CD163⁺ M2 macrophages.
.jpg) Figure 2. Single-cell transcriptomic profiling. A) Ten distinct cell clusters were delineated. Black cluster corresponded to CD163-positive M2 macrophages. B, C) Molecular network construction and topological analysis of the top 30 differentially expressed genes in the CD163-positive M2 macrophage cluster identified two hub genes.
Figure 2. Single-cell transcriptomic profiling. A) Ten distinct cell clusters were delineated. Black cluster corresponded to CD163-positive M2 macrophages. B, C) Molecular network construction and topological analysis of the top 30 differentially expressed genes in the CD163-positive M2 macrophage cluster identified two hub genes.
.jpg) Figure 3. Comparison of the expression of two identified hub genes in muscle biopsy specimens from five healthy subjects and five patients with dermatomyositis. All identified hub genes exhibited a trend toward elevated expression in dermatomyositis patients.
Figure 3. Comparison of the expression of two identified hub genes in muscle biopsy specimens from five healthy subjects and five patients with dermatomyositis. All identified hub genes exhibited a trend toward elevated expression in dermatomyositis patients.
To cite this abstract in AMA style:
Harada T, Yamashita H, Isoda A, Kawaue K, Hayashi M, Misawa Y, Yamamoto A, Wakatsuki M, Akiyama Y, Oyama S, Motomura K, Takahashi H, Mitsuo A, Goto Y, Noiri E, Kaneko H. Single-cell and Spatial Transcriptomic Profiling of Muscle Reveals Inflammatory Mechanisms in Anti-glycyl tRNA Synthetase Syndrome [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/single-cell-and-spatial-transcriptomic-profiling-of-muscle-reveals-inflammatory-mechanisms-in-anti-glycyl-trna-synthetase-syndrome/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/single-cell-and-spatial-transcriptomic-profiling-of-muscle-reveals-inflammatory-mechanisms-in-anti-glycyl-trna-synthetase-syndrome/
