Session Information
Date: Sunday, November 17, 2024
Title: RA – Treatment Poster II
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: Our study aimed to use machine-learning approaches to characterize the proteomics profiles of patients who were inadequate responders to Etanercept (ETN-IRs) and develop an ETN-IR risk model based on proteomic signatures in rheumatoid arthritis (RA).
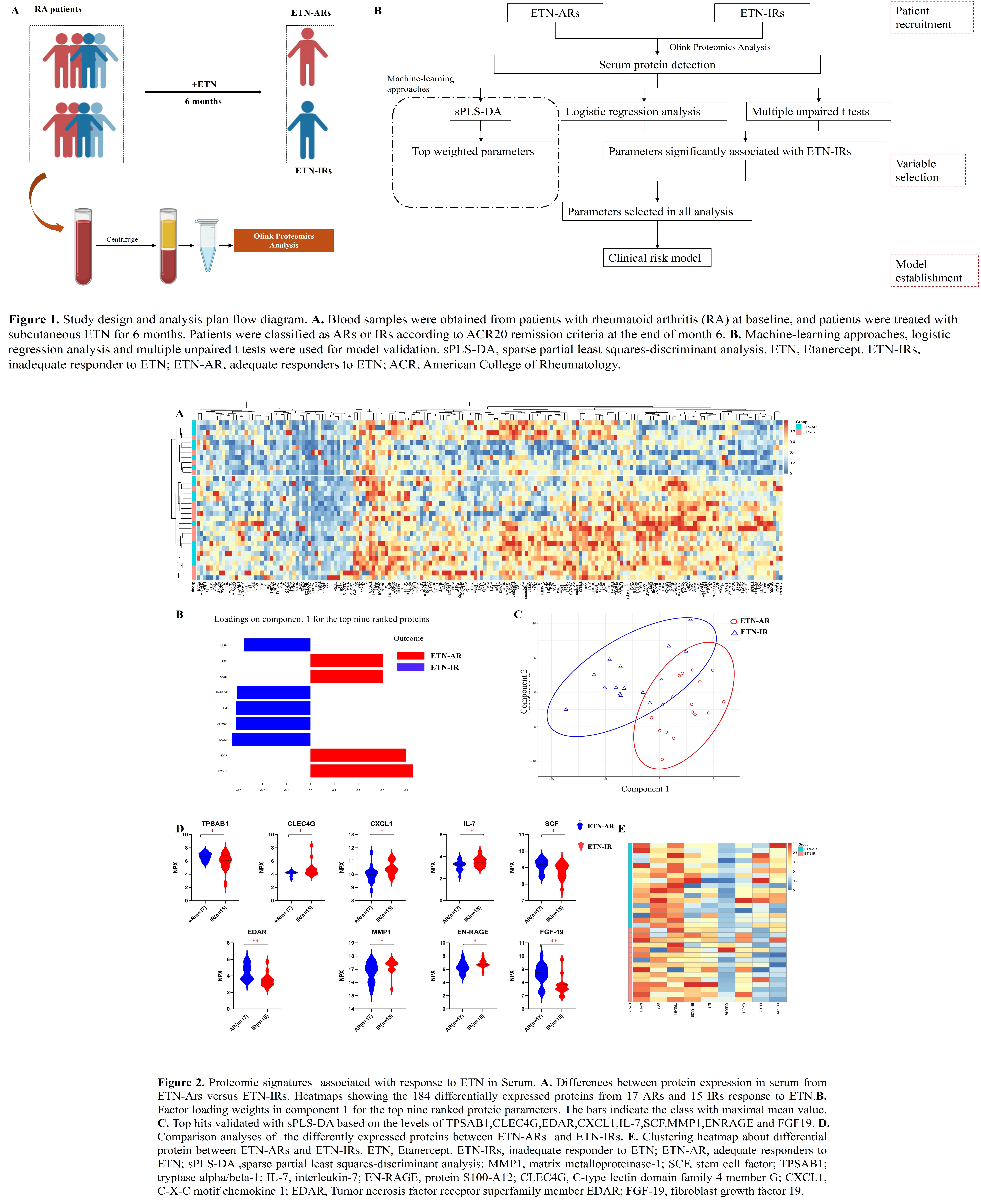
Methods: The study included 32 RA patients from the Department of Rheumatology and Immunology, Peking University People’s Hospital between July, 2022 and November, 2023. All the patients fulfilled the 2010 ACR and EULAR classification criteria for RA and received ETN 25mg twice a week for 6 months. Patients who didn’t achieve ACR20 improvement at 6th month after enrollment were defined as ETN-IRs and others were defined as ETN adequate responders (ETN-ARs). One hundred eighty-four serum proteins from ETN-IRs and ETN-ARs were detected by Olink Proteomics Analysis. Sparse partial least squares-discriminant analysis (sPLS-DA), univariate and multivariate logistic regression analyses were performed to identify proteomic signatures associated with ETN-IRs to build a risk model. Study design and analysis plan flow diagram was shown in figure1.
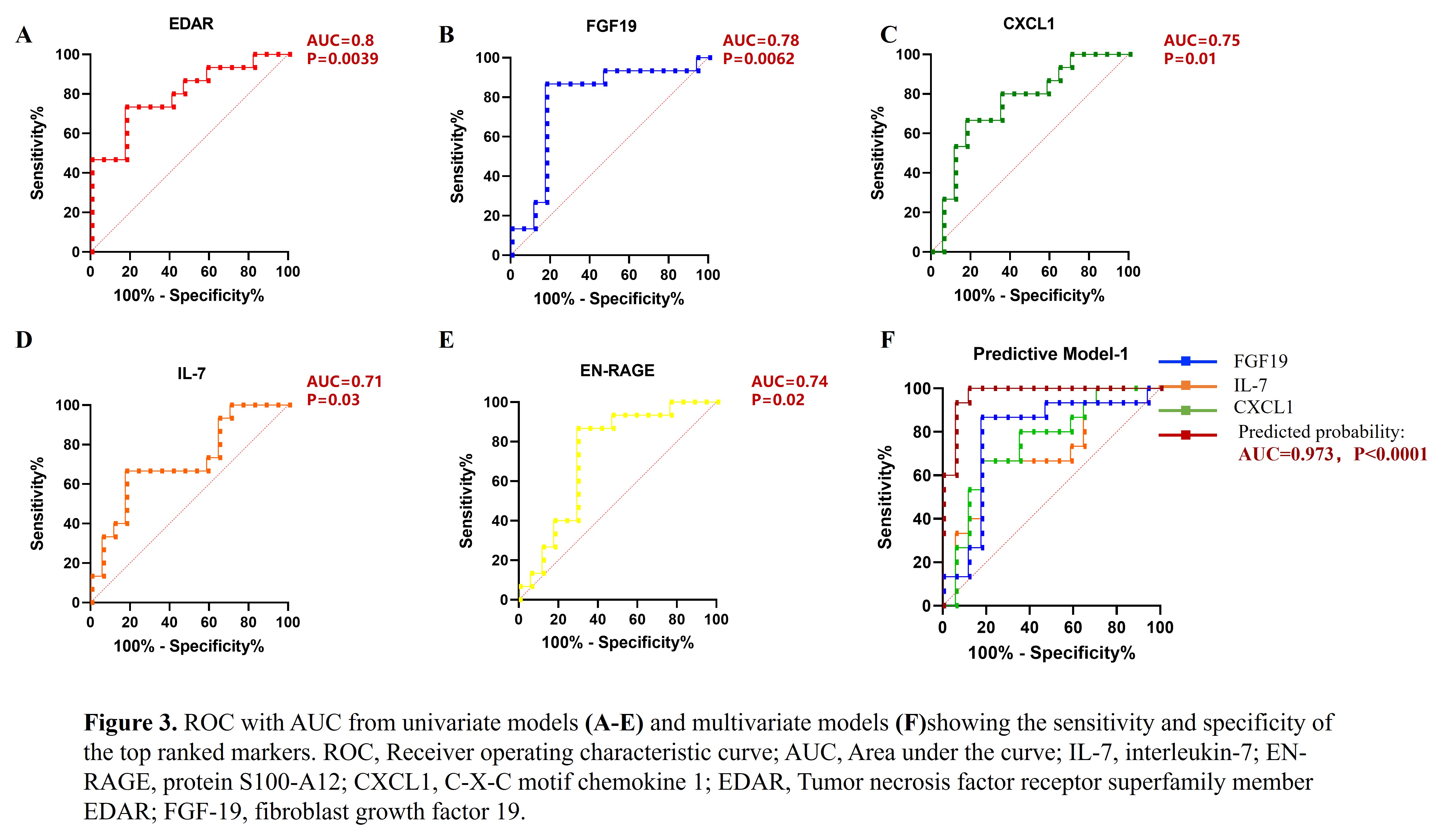
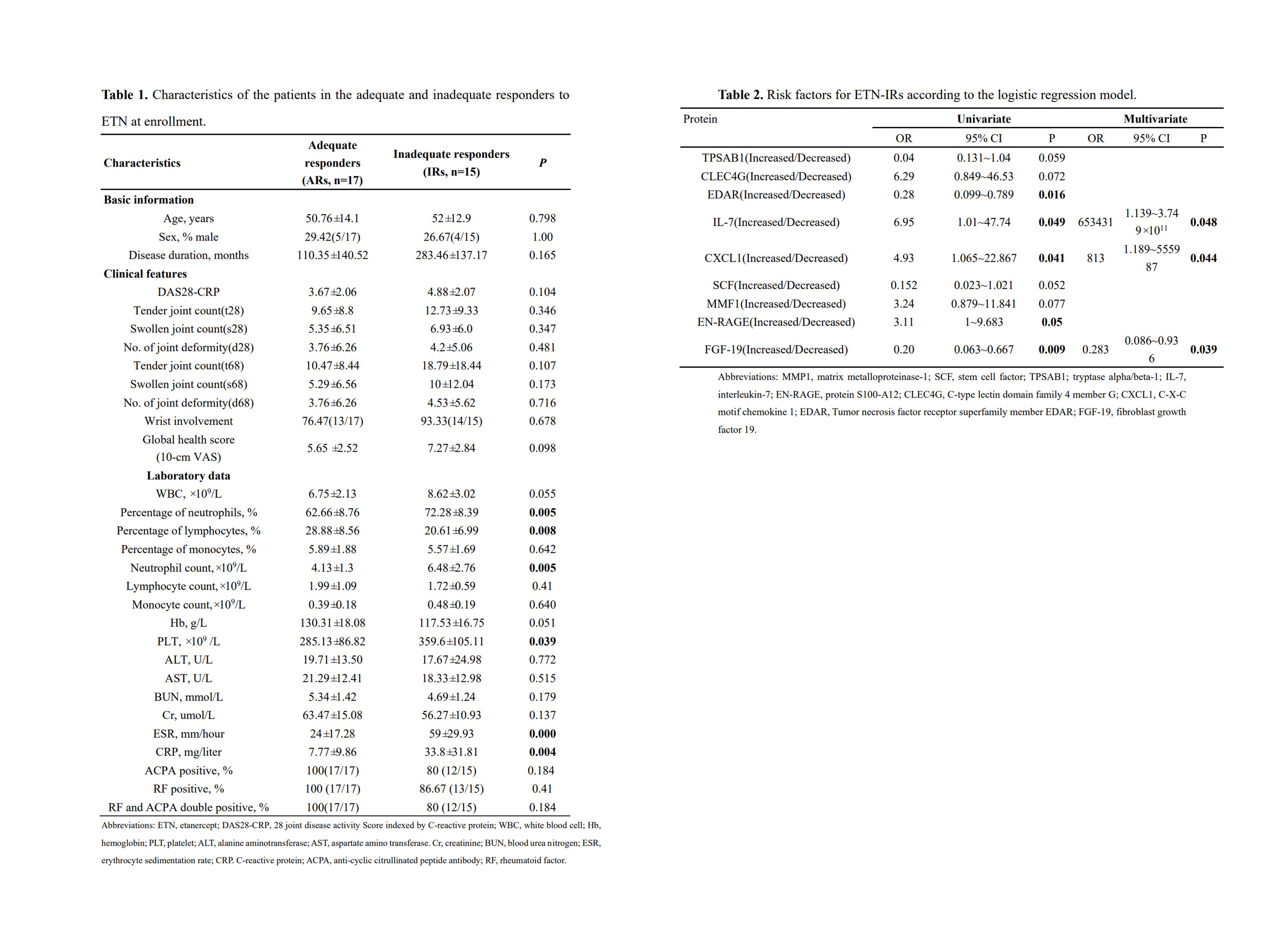
Results: The demographic data and clinical features of the patients were described in table 1. We first studied the proteomic signatures in serum and identified 184 differentially expressed proteins between ETN-ARs and ETN-IRs (figure 2A). Factor loading weights in component 1 for the top nine ranked proteomic parameters were shown in figure 2B. To validate the relationship of the top nine proteomic variables and response to ETN, sPLSDA was done and identified a significant separation between ETN-ARs and ETN-IRs by plotting principal component 2 against principal component 1(figure 2C).In-depth proteomic analysis showed that ETN-IRs had a disrupted proteomic profile compared with ETN-ARs, including alteration in serum levels of MMP1, SCF, TPSAB1, IL-7, EN-RAGE, CLEC4G, CXCL1, EDAR and FGF-19 (figure 2D-E). We identified that the increased levels of IL-7 and CXCL1 but decreased levels of FGF-19 in serum were the independent risk factors for ETN-IRs (table 2). Area under the curve (AUC) from univariate models showing the sensitivity and specificity of the top five markers identified by the model (figure3A-E). A multi-parametric prediction model for ETN-IRs was established based on weighted proteomic signatures including FGF-19, IL-7and CXCL1. Receiver operating characteristic curve (ROC) analysis of the model showed an AUC of 0.973 (sensitivity 93.3%, specificity 94.6%) (figure 3F), indicating good performance in discriminating ETN-IRs from ETN-ARs.
Conclusion: Our findings indicate that the models based on proteomic signatures accurately predict response before ETN treatment, paving the path toward personalized anti-TNF treatment.
Figure 2. Proteomic signatures associated with response to ETN in Serum. A. Differences between protein expression in serum from ETN-Ars versus ETN-IRs. Heatmaps showing the 184 differentially expressed proteins from 17 ARs and 15 IRs response to ETN.B. Factor loading weights in component 1 for the top nine ranked proteic parameters. The bars indicate the class with maximal mean value. C. Top hits validated with sPLS-DA based on the levels of TPSAB1, CLEC4G, EDAR, CXCL1, IL-7, SCF, MMP1, ENRAGE and FGF19. D. Comparison analyses of the differently expressed serum proteins between ETN-ARs and ETN-IRs. E. Clustering heatmap about differential protein between ETN-ARs and ETN-IRs. ETN, Etanercept. ETN-IRs, inadequate responder to ETN; ETN-AR, adequate responders to ETN; sPLS-DA ,sparse partial least squares-discriminant analysis; MMP1, matrix metalloproteinase_1; SCF, stem cell factor; TPSAB1; tryptase alpha/beta_1; IL-7, interleukin-7; EN-RAGE, protein S100-A12; CLEC4G, C-type lectin domain family 4 member G; CXCL1, C-X-C motif chemokine 1; EDAR, Tumor necrosis factor receptor superfamily member EDAR; FGF_19, fibroblast growth factor 19.
Table 2. Risk factors for ETN-IRs according to the logistic regression model.
To cite this abstract in AMA style:
Zhu H, cheng G, Li y, Li Y, Sun F, Wang H, li Q, Li Z, Li R, Li Z. Proteomics and Machine Learning Accurately Predict Clinical Response to Etanercept Therapy in Patients with Rheumatoid Arthritis [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/proteomics-and-machine-learning-accurately-predict-clinical-response-to-etanercept-therapy-in-patients-with-rheumatoid-arthritis/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/proteomics-and-machine-learning-accurately-predict-clinical-response-to-etanercept-therapy-in-patients-with-rheumatoid-arthritis/