Session Information
Date: Sunday, November 12, 2023
Title: (0252–0282) Miscellaneous Rheumatic & Inflammatory Diseases Poster I
Session Type: Poster Session A
Session Time: 9:00AM-11:00AM
Background/Purpose: Autoinflammatory diseases (AID) are characterized by inflammation and immunopathology due to primary defects in the innate immune response. Neutrophils (PMN) feature prominently in the biology of most AID, but show clear differences between AID categories (e.g. mature PMN in NLRP3-AID, atypical PMN in proteasome-AID, low-density granulocytes in interferonopathies). However, PMN are excluded in mononuclear cell preparations (e.g. PBMC), do not tolerate freeze/thaw, have low mRNA content, and change dramatically & quickly with treatment. Thus, optimal neutrophil studies require rapid preservation of high-quality samples from unpredictable “flare” timepoints prior to significant pharmacologic intervention. Fixed-cell Indexing of Transcriptomes and Epitopes sequencing (FITEseq) is a novel method for quickly processing and preserving epitope and transcript abundance. We sought to understand which cells were excluded from PBMC preparations, and to pilot the feasibility and quality of whole blood FITEseq in active AID patients.
Methods: Pellets from patient PBMC preparations underwent magnetic red blood cell (RBC) depletion and flow cytometry. 1mL of K-EDTA anticoagulated whole blood from a new-onset Stills-like patient and one healthy control were RBC depleted, fixed (10x Single Cell Fixed RNA Sample Preparation Kit), and stored at -80C. Parallel samples were analyzed by flow cytometry. Libraries prepared using the Chromium human fixed RNA kit, RNA integrity determined by bioanalyzer, libraries quantitated by Qbit and sequenced on NextSeq1000 (P2 reagents, 100 cycles). Analysis was performed using CellRanger7 and visualizations performed using Cellenics.
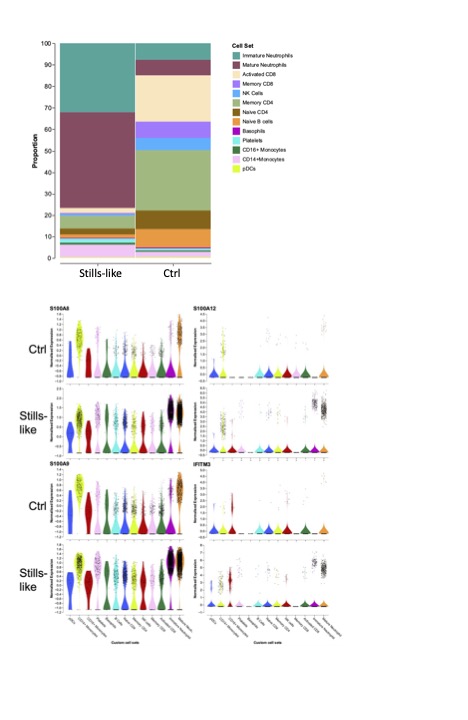
Results: In addition to CD15+ neutrophils, PBMC pellets variably included monocyte populations, predominantly in patient samples, that would not be included in buffy coats. Whole blood preparation resulted in minimal death or specific loss of neutrophils. Preliminary technical analyses highlighted the importance of timely processing, precise library quantitation, and low-RNA permissive analysis. Contemporaneous Stills-like patient serum showed dramatic total IL-18 and S100A12 elevation. The proportion of neutrophils identified transcriptionally matched well with contemporaneous differential, and was higher in the patient (Fig. 1). Exploratory analysis showed higher expression of granule, S100, and interferon-inducible genes in patient myeloid cells, and CD38 on patient CD8 T-cells.
Conclusion: Single-cell analysis of fixed whole blood from AID patients is feasible and demonstrates physiologically-relevant alterations in cells omitted in PBMC preparations (including PMN and monocyte populations). FITEseq may be a practical and scalable method of obtaining single-cell protein and transcriptome data from samples containing fragile myeloid cells.
To cite this abstract in AMA style:
Carol H, Landy E, Canna S. Preliminary Experience with a Novel “Fix” for Deep Epitope and Transcriptional Phenotyping of Fragile Cells from Autoinflammatory Flares [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/preliminary-experience-with-a-novel-fix-for-deep-epitope-and-transcriptional-phenotyping-of-fragile-cells-from-autoinflammatory-flares/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/preliminary-experience-with-a-novel-fix-for-deep-epitope-and-transcriptional-phenotyping-of-fragile-cells-from-autoinflammatory-flares/