Session Information
Session Type: ACR Poster Session B
Session Time: 9:00AM-11:00AM
Background/Purpose: Phenotype identification in knee osteoarthritis (KOA) population could be useful for predicting drug response, allowing personalized interventions. In order to optimize therapeutic outcome in KOA, we aimed to predict drug response of patients treated with COX-2 selective nonsteroidal anti-inflammatory drug Celecoxib (CLX) or pharmaceutical grade Chondroitin sulfate plus glucosamine hydrochloride (CS+GH) combining the analysis of multiple clinical variables and omics data.
Methods: A shotgun proteomic analysis by iTRAQ was performed on sera from 80 patients enrolled in the Multicentre Osteoarthritis interVEntion trial with Sysadoa (MOVES). Then, a panel of 10 serum proteins was qualified using ELISA Kits in the whole MOVES cohort (n=1043). Patients were classified as responders (R) and non-responders (NR), either to CLX or CS+GH according to the OMERACT-OARSI criteria and the WOMAC pain score recorded after 6 months of treatment. Logistic regression analyses, adjusted by significant confounder variables, were used to analyze the contribution of the measured proteins to our prediction models of drug response in KOA. Appropriate receiver-operating-characteristics (ROC) curves were also calculated.
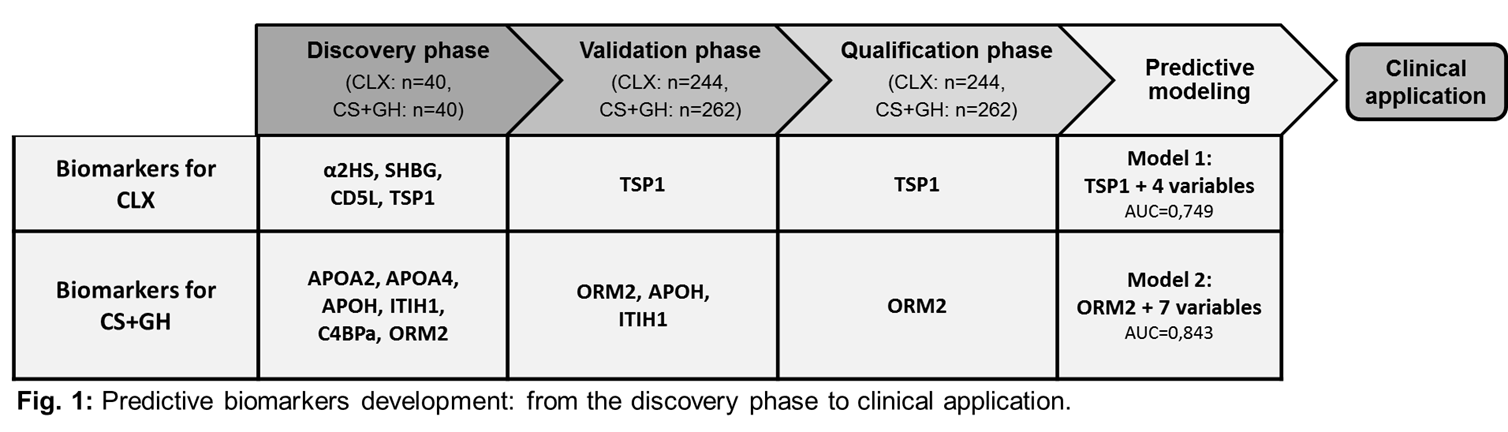
Results: In the discovery phase of the study, the proteomic screening led to the identification of 83 proteins significantly altered at baseline in R compared to NR. Among the proteins presenting the highest iTRAQ ratios and exclusively altered in one of the therapeutic groups, we selected 4 proteins specific for CLX treatment and 6 proteins specific for CS+GH treatment for the development of the validation assays in a larger cohort of KOA patients (Fig. 1). In the qualification phase, the sensitivity and specificity of the validated proteins were tested in blind in the whole MOVES cohort at baseline. In the CLX group, an increased level of TSP1 was detected at baseline in R compared to NR (363,03 ng/mL vs 331,95 ng/mL; p=0,041). The inclusion in the regression model of 4 predictive variables (2 clinical and 2 analytical) and TSP1 as covariate revealed a specific interaction between response to CLX and baseline protein levels (p=0,045) thus increasing the predictive power of this model up to AUC=0,749 (Model 1, Fig. 1). In CS+GH group, ORM2 levels were significantly higher in NR compared to R (261,6 ug/mL vs 192,8 ug/mL; p=0,042). 5 clinical and 2 analytical parameters recorded at baseline significantly influence patients’ response. The inclusion of ORM2 as covariate revealed a specific interaction between response to CS+GH and baseline protein levels (p=0,007) thus increasing the power of our prediction model up to AUC=0,843 (Model 2, Fig.1).
Conclusion: Combining clinical and analytical parameters, we qualified 2 panels of biomarkers that could efficiently predict OA patients’ response to CLX with an accuracy of 74,9% or to CS+GH with an accuracy of 84,3%.
To cite this abstract in AMA style:
Calamia V, Picchi F, Rego-Pérez I, Camacho M, González L, Fernández P, Herrero M, Martinez H, Vergés J, Ruiz-Romero C, Blanco FJ. Predictive Modeling of Therapeutic Response in Knee Osteoarthritis [abstract]. Arthritis Rheumatol. 2018; 70 (suppl 9). https://acrabstracts.org/abstract/predictive-modeling-of-therapeutic-response-in-knee-osteoarthritis/. Accessed .« Back to 2018 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/predictive-modeling-of-therapeutic-response-in-knee-osteoarthritis/