Session Information
Date: Sunday, October 26, 2025
Session Type: Abstract Session
Session Time: 3:30PM-3:45PM
Background/Purpose: Lupus nephritis (LN), a severe complication of systemic lupus erythematosus (SLE), lacks reliable non-invasive biomarkers for early detection and monitoring. This study investigates plasma cell-free DNA (cfDNA) methylation and fragmentation signatures as potential LN biomarkers.
Methods: We performed whole-genome methylation profiling of plasma cfDNA from distinct cohorts: SLE patients with biopsy-proven LN (active: n=10 and inremission: n=7), SLE patients without LN ( >10 years post-diagnosis, n=13), and healthy controls (n=10). Two novel methylation-based deconvolution methods were developed to quantify kidney-derived cfDNA and compared to classical non-negative least square (NNLS)algorithms. Fragmentation patterns were analyzed using cfDNA size profiles to differentiate active LN from remission.
Results: Our novel deconvolution methods outperformed NNLS in detecting low-abundance cell types. Active LN patients exhibited significantly elevated kidney-derived cfDNA levels compared to remission, never-LN SLE, and healthy cohorts (p< 0.05 for all) (Figure 1). The reads ratio-based method achieved high diagnostic accuracy for active LN (AUC=0.91) (Figure 2). All three methods strongly correlated kidney-derived cfDNA levels with LN disease activity. Active LN patients also exhibited increased short cfDNA fragments (30–157 bp) compared to remission (p< 0.001) (Figure 3). Combining NNLS-derived kidney cfDNA levels with short fragment proportions enabled exceptional discrimination between active and remission LN (AUC=1).
Conclusion: Kidney-derived cfDNA levels and short fragment proportions are robust biomarkers for distinguishing active LN from remission and never-LN SLE. These findings underscore the clinical potential of cfDNA methylation and fragmentation profiling as a non-invasive tool for LN diagnosis, monitoring, and personalized treatment strategies.
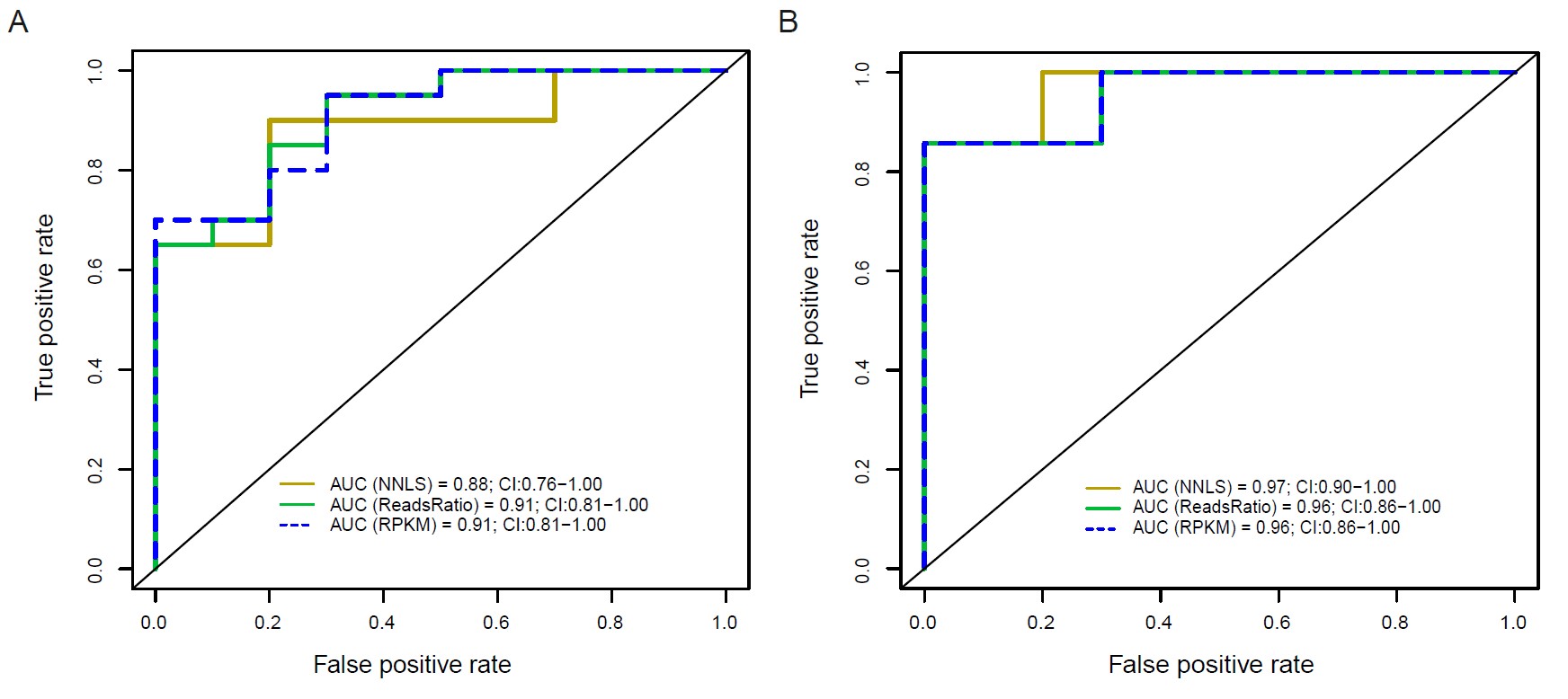 Figure 1. Kidney-derived cfDNA levels measured by three deconvolution methods are significantly elevated in active LN patients. *p < 0.05, **p < 0.01, ***p < 0.001.
Figure 1. Kidney-derived cfDNA levels measured by three deconvolution methods are significantly elevated in active LN patients. *p < 0.05, **p < 0.01, ***p < 0.001.
.jpg) Figure 2. Receiver operating characteristic (ROC) curves of kidney-derived cfDNA levels profiled by NNLS, ReadsRatio and RPKM deconvolution
Figure 2. Receiver operating characteristic (ROC) curves of kidney-derived cfDNA levels profiled by NNLS, ReadsRatio and RPKM deconvolution
algorithms distinguishing SLE patients with active LN from all other SLE patients (A) and SLE patients with active LN from those with LN in remission (B).
.jpg) Figure 3. cfDNA fragmentation pattern for identification of active LN patients (red) and inactive LN patients (Grey). Plasma cfDNA fragments of 30-157bp are enriched in active LN patients compared to inactive LN patients. ***p < 0.001.
Figure 3. cfDNA fragmentation pattern for identification of active LN patients (red) and inactive LN patients (Grey). Plasma cfDNA fragments of 30-157bp are enriched in active LN patients compared to inactive LN patients. ***p < 0.001.
To cite this abstract in AMA style:
Jing Q, Li Q, Chan C, Chen W, choi F, Tang I, Lau C, Ling H, Wong J, Li P. Precision Liquid Biopsy for Lupus Nephritis: cfDNA Methylation and Fragmentomic Signatures Enable Non-Invasive Diagnosis and Dynamic Disease Tracking [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/precision-liquid-biopsy-for-lupus-nephritis-cfdna-methylation-and-fragmentomic-signatures-enable-non-invasive-diagnosis-and-dynamic-disease-tracking/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/precision-liquid-biopsy-for-lupus-nephritis-cfdna-methylation-and-fragmentomic-signatures-enable-non-invasive-diagnosis-and-dynamic-disease-tracking/
