Session Information
Date: Monday, November 11, 2019
Title: Systemic Sclerosis & Related Disorders – Basic Science Poster
Session Type: Poster Session (Monday)
Session Time: 9:00AM-11:00AM
Background/Purpose: We have examined whole skin biopsy gene expression by RNAseq in a rare subgroup of scleroderma with both systemic sclerosis (SSc) and concurrent keloidal morphea (KM). We hypothesised that this subtype of localised scleroderma would provide exceptional insight into fibroblast activation in vivo relevant to skin fibrosis in systemic sclerosis and suggest new potential molecular markers for classification.
Methods: 4mm skin biopsies were taken from forearm skin of SSc cases classified as limited (lcSSc) (n=5) or diffuse (dcSSc) (n=7). All patients met ACR/EULAR criteria for classification of SSc. In 4 cases there was concurrent KM and 4mm skin were also taken from these lesions. Control biopsies were taken from forearm skin of healthy individuals (HC) (n=4). Whole genome expression analysis was performed by RNAseq. Data were normalised and scaled. Gene expression analysis was undertaken and differentially expressed genes were compared across the clinical subgroups using ANOVA with Benjamini-Hochberg post-hoc correction. The 500 most differentially expressed genes were identified, and unsupervised clustering was performed using CIMminer (Bethesda, Maryland, USA). Integration of the candidate genes from ANOVA, and principal component analysis (PCA) was carried out to identify instructive genes for a molecular classifier.
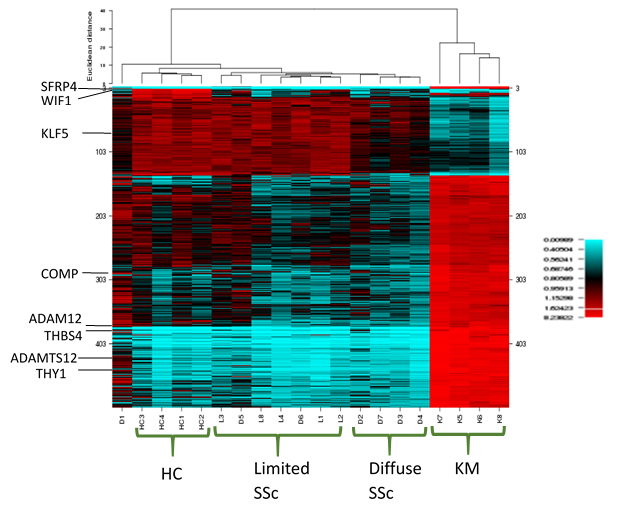
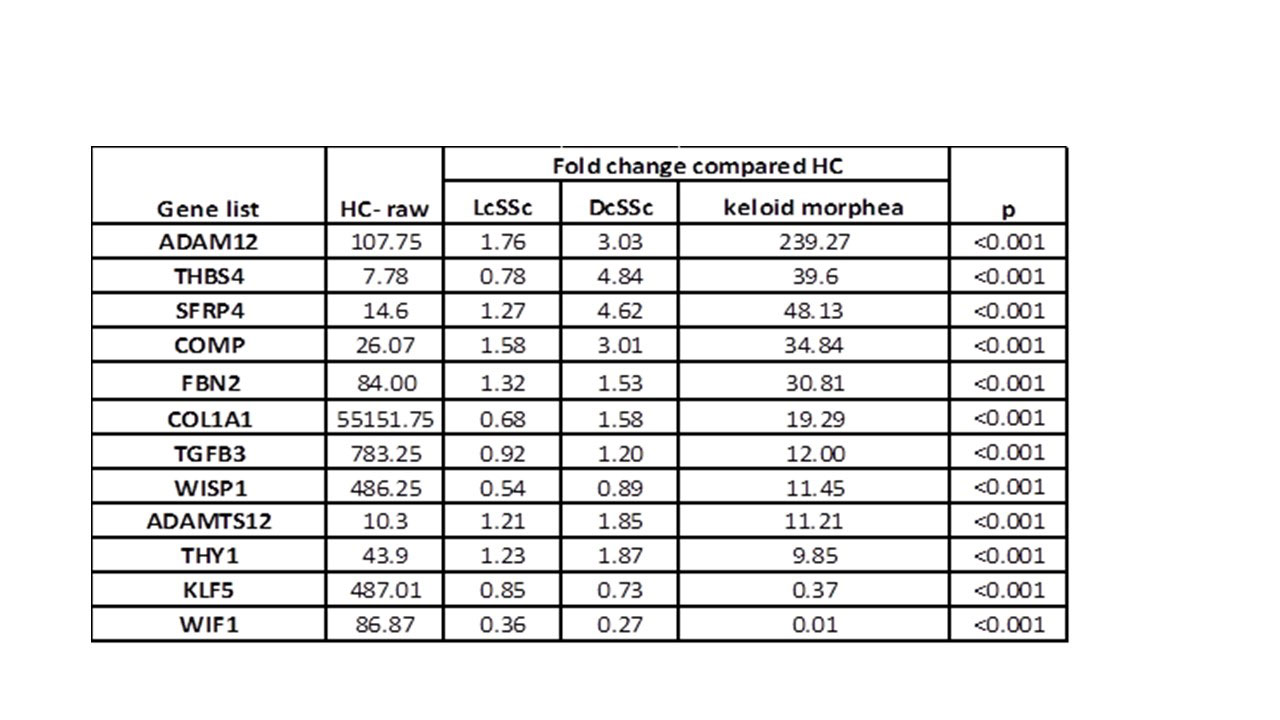
Results: RNAseq identified over 13000 expressed genes. PCA discriminated 4 unique clusters, with keloidal morphea and HCs being the most distinct. PC1 accounted for 33% and PC2 for 22% of variation. Initial analysis identified over 3000 significantly different genes expressed by paired analysis. The 500 most significantly differentially expressed genes (all with p< 0.001) were selected. Unsupervised hierarchical clustering of gene expression based on these results, showed clear clustering of the keloidal morphea group, and the HCs. The majority of lcSSc and dcSSc patients clustered to their patient subgroups (Figure 1). Correlation between ANOVA and PCA results highlighted 100 key genes shared across both analysis results. These included genes upregulated in SSc: SFRP4, THY1, COMP, ADAM12, THBS4, ADAMTS12 (table 1) and others with lower expression than in HC: WIF1, KLF5. These include several implicated in pathogenesis or included in recent candidate biomarkers for skin disease in SSc.
Conclusion: We show the high value of RNAseq and the unique strength using skin biopsies from SSc with concurrent keloidal morphea, histologically characterised by dense fibro-proliferation, to define profibrotic genes relevant to SSc. This provides powerful insight into pathogenesis and candidate molecular markers for classification across the scleroderma spectrum.
To cite this abstract in AMA style:
Clark K, Campochiaro C, Gak N, Derrett-Smith E, Denton C. Parallel Analysis of Systemic Sclerosis and Keloidal Morphea Skin Biopsies Delineates the Hallmark Profibrotic Gene Expression Profile for Scleroderma in Vivo [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/parallel-analysis-of-systemic-sclerosis-and-keloidal-morphea-skin-biopsies-delineates-the-hallmark-profibrotic-gene-expression-profile-for-scleroderma-in-vivo/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/parallel-analysis-of-systemic-sclerosis-and-keloidal-morphea-skin-biopsies-delineates-the-hallmark-profibrotic-gene-expression-profile-for-scleroderma-in-vivo/