Session Information
Date: Monday, November 9, 2020
Title: Pediatric Rheumatology – Clinical III: Systemic Autoimmune Disease (1988–1992)
Session Type: Abstract Session
Session Time: 11:00AM-11:50AM
Background/Purpose: Skin inflammation can herald systemic disease in juvenile myositis (JM), yet we lack an understanding of pathogenic mechanisms driving skin inflammation in JM. The goals of this study were to 1) define JM cutaneous gene expression signatures in the context of patient data, and 2) identify key genes and pathways differentiating skin disease in JM from childhood-onset systemic lupus erythematosus (cSLE).
Methods: We utilized formalin-fixed paraffin-embedded (FFPE) skin biopsies from 15 JM (9 lesional, 6 non-lesional), 5 cSLE (all lesional), and 8 controls to perform transcriptomic analysis and identify significantly differentially expressed genes (DEGs; q-value ≤ 5%) between patient groups. Ingenuity Pathway Analysis (IPA) was used to highlight enriched biological pathways and Genomatix Pathway System (GePS) to characterize regulated genes within biological networks. We validated DEGs using immunohistochemistry and quantitative real-time PCR. Interferon (IFN) scores were calculated and compared amongst patient groups and between JM clinical features. A JM-specific signature was derived from comparison of cSLE and lesional JM (JM_L) skin transcriptomes relative to controls and compared to adult skin disease array datasets. Cell type enrichment analysis was then performed using the xCell webtool.
Results: Comparison of JM_L to control revealed 221 DEGs, with the majority of upregulated genes representing IFN-stimulated genes. CXCL10, CXCL9 and IFI44L represented the top three DEGs (fold-change respectively = 23.2, 13.3, 13.0, q-value < 0.0001). While IFN scores in JM did not differ based on individual skin disease manifestations or treatment status, NXP2+ JM patients exhibited the strongest IFN signature (Figure 1) and also demonstrated the most extensive MX1 immunostaining, both in keratinocytes and perivascular regions. When compared to cSLE, JM_L skin showed no difference in IFN scores and shared a similar gene expression pattern, with only 28 unique DEGs. The top most significant unique DEGs in JM_L included FBLN2, CHKA and SLURP1, genes with diverse roles in extracellular matrix structure, keratinocyte proliferation and differentiation, calcium signaling and phospholipid metabolism. A 3-gene JM-specific skin signature derived using FBLN2, CHKA and SLURP1 was higher in dermatomyositis (both pediatric and adult) as compared to other skin diseases, including cutaneous lupus (Figure 2). cSLE skin had 722 unique DEGs compared to JM_L, notably increased expression of IFNγ relative to control. Cell type enrichment analysis showed that cSLE skin exhibited an overall higher inflammatory cell signature compared to JM_L, with increased T-cells, B-cells, macrophages and plasmacytoid dendritic cells (Figure 3).
Conclusion: JM lesional skin demonstrates a prominent IFN signature, similar to cSLE. A candidate JM-specific skin signature was derived using FBLN2, CHKA and SLURP1, all genes not typically considered to have immunomodulatory roles but instead functions in cellular structure and metabolism. Further investigation into the association of a higher IFN score with NXP2 autoantibodies may lend insight into JM disease endotypes and pathogenesis.
 Figure 1. JM IFN score with clinical variables. A. The IFN score is not significantly modified by the presence of systemic disease, nailfold capillary changes or calcinosis. B. The IFN score is not significantly changed by treatment status. C. The presence alone of a myositis-specific autoantibody (MSA) does not significantly alter the IFN score; however, NXP2+ JM patients have a significantly higher IFN score (p-value=0.034). D. An increased overall number of serum muscle enzymes was associated with a higher IFN score (p-value=0.0381). A higher IFN score demonstrated a trend toward presence of dysphagia.
Figure 1. JM IFN score with clinical variables. A. The IFN score is not significantly modified by the presence of systemic disease, nailfold capillary changes or calcinosis. B. The IFN score is not significantly changed by treatment status. C. The presence alone of a myositis-specific autoantibody (MSA) does not significantly alter the IFN score; however, NXP2+ JM patients have a significantly higher IFN score (p-value=0.034). D. An increased overall number of serum muscle enzymes was associated with a higher IFN score (p-value=0.0381). A higher IFN score demonstrated a trend toward presence of dysphagia.
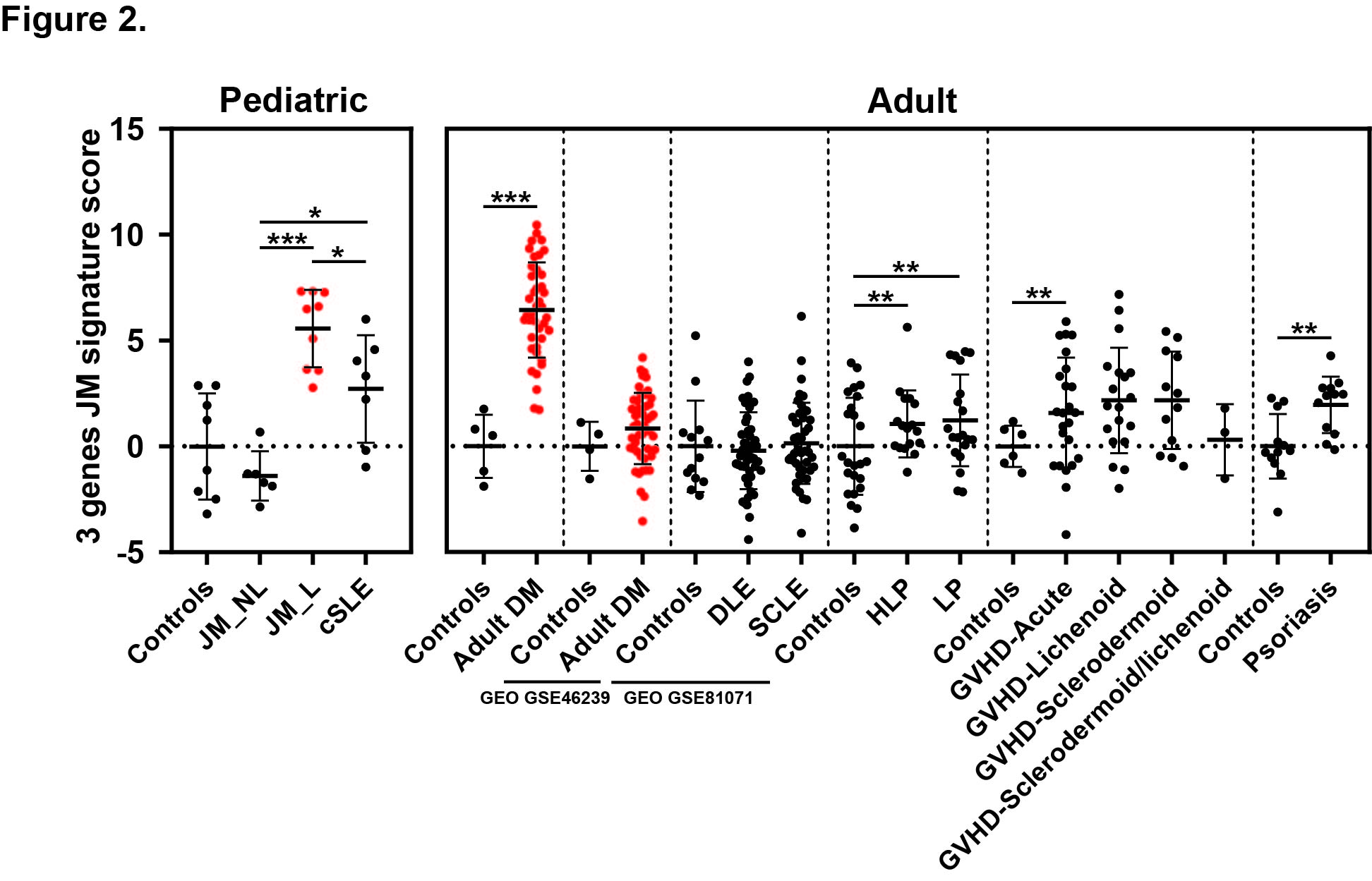 Figure 2. JM disease signature: comparison with transcriptomic datasets of skin lesions from adult DM and other inflammatory skin diseases. The 3-gene JM transcriptomic signature identified is the highest in juvenile and adult dermatomyositis compared to other skin disease lesions. Dermatomyositis lesional samples are represented in red. Vertical dashed lines separate the studied datasets. Each dataset had its control sample set. DLE: discoid lupus erythematosus, SCLE: subacute cutaneous lupus erythematosus, HLP: Hypertrophic lichen planus, LP: lichen planus, GVHD: graft versus host disease. * p-value < 0.05; ** p-value < 0.01; *** p-value < 0.0001.
Figure 2. JM disease signature: comparison with transcriptomic datasets of skin lesions from adult DM and other inflammatory skin diseases. The 3-gene JM transcriptomic signature identified is the highest in juvenile and adult dermatomyositis compared to other skin disease lesions. Dermatomyositis lesional samples are represented in red. Vertical dashed lines separate the studied datasets. Each dataset had its control sample set. DLE: discoid lupus erythematosus, SCLE: subacute cutaneous lupus erythematosus, HLP: Hypertrophic lichen planus, LP: lichen planus, GVHD: graft versus host disease. * p-value < 0.05; ** p-value < 0.01; *** p-value < 0.0001.
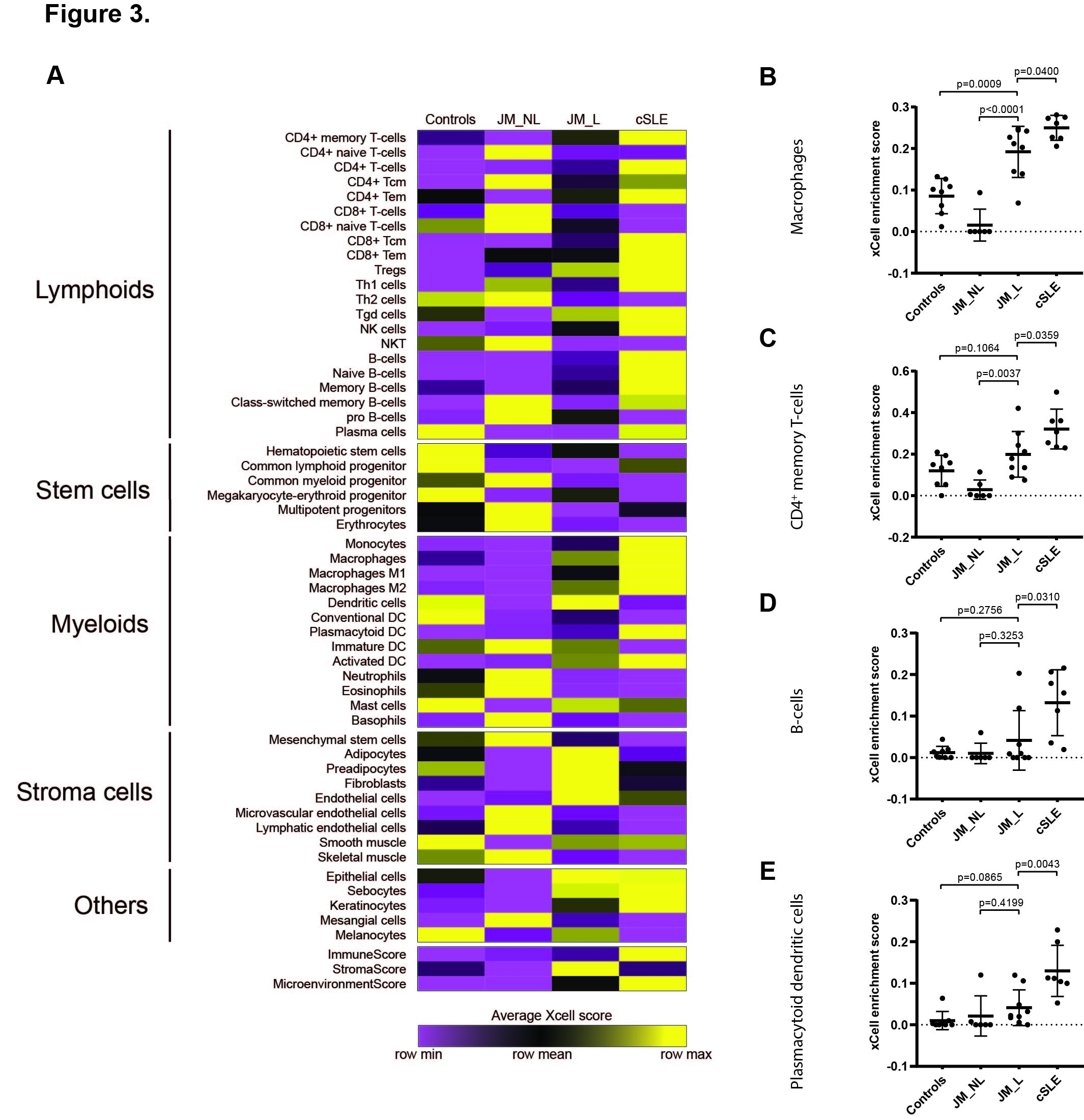 Figure 3. Cell type enrichment analysis using xCell webtool. A. Heatmap of relevant cell types representing average xCell enrichment score for controls, JM_NL, JM_L and cSLE. B. Graph of the macrophage xCell score, showing a significantly higher score in JM_L compared to JM_NL and controls and in cSLE compared to JM_L. C. Graph of the CD4+ memory T-cells xCell score, showing a significantly higher score in JM_L compared to JM_NL and in cSLE compared to JM_L. D. Graph of the B-cells xCell score showing a significantly higher score in cSLE compared to JM_L. E. Graph of the plasmacytoid dendritic cells xCell score, showing a significantly higher score in cSLE compared to JM_L. For (B-E) comparisons were made via unpaired Students’ t-test.
Figure 3. Cell type enrichment analysis using xCell webtool. A. Heatmap of relevant cell types representing average xCell enrichment score for controls, JM_NL, JM_L and cSLE. B. Graph of the macrophage xCell score, showing a significantly higher score in JM_L compared to JM_NL and controls and in cSLE compared to JM_L. C. Graph of the CD4+ memory T-cells xCell score, showing a significantly higher score in JM_L compared to JM_NL and in cSLE compared to JM_L. D. Graph of the B-cells xCell score showing a significantly higher score in cSLE compared to JM_L. E. Graph of the plasmacytoid dendritic cells xCell score, showing a significantly higher score in cSLE compared to JM_L. For (B-E) comparisons were made via unpaired Students’ t-test.
To cite this abstract in AMA style:
Turnier J, Pachman L, Lowe L, Tsoi A, Elhaj S, Menon R, Amoruso M, Morgan G, Gudjonsson J, Berthier C, Kahlenberg J. NXP2 Autoantibodies Link to Interferon Signature in Juvenile Myositis Lesional Skin [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/nxp2-autoantibodies-link-to-interferon-signature-in-juvenile-myositis-lesional-skin/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/nxp2-autoantibodies-link-to-interferon-signature-in-juvenile-myositis-lesional-skin/
