Session Information
Session Type: Abstract Session
Session Time: 2:45PM-3:00PM
Background/Purpose: The clinical heterogeneity of SLE with its complex pathogenesis remains challenging as we strive to provide optimal management. The contribution of platelets to endovascular homeostasis, inflammation and immune regulation highlights their potential importance in SLE. Prior work from our group showed that the Fcγ receptor type IIa (FcγRIIa)–R/H131 biallelic polymorphism is associated with increased platelet activity and cardiovascular risk in SLE. The study was initiated to investigate the platelet transcriptome in patients with SLE and evaluate its association across FcγRIIa genotypes and distinct clinical features.
Methods: RNA-sequencing was done on platelets isolated from 51 patients fulfilling criteria for the classification of SLE based on recent EULAR/ACR definitions, and 18 healthy controls matched on age, sex, and race. Unsupervised clustering, differential gene expression, and gene set enrichment analysis (GSEA) were used to analyze differences between SLE patients and controls, and SLE subpopulations, based on SELENA SLEDAI Hybrid disease activity, specific organ manifestations, and FcγRIIa genotype. Weighted Gene Correlation Network Analysis (WGCNA) was performed to create a modular transcriptomic framework.
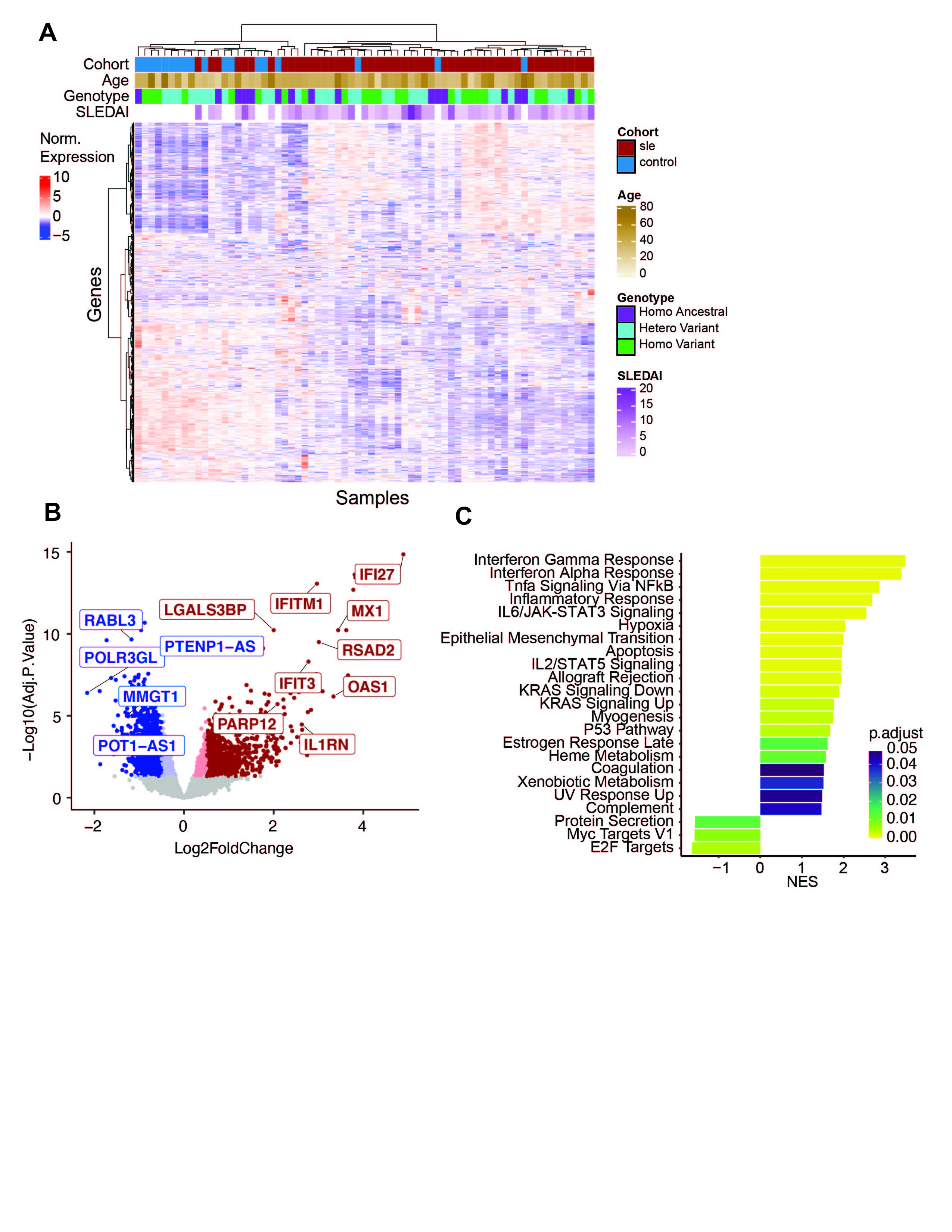
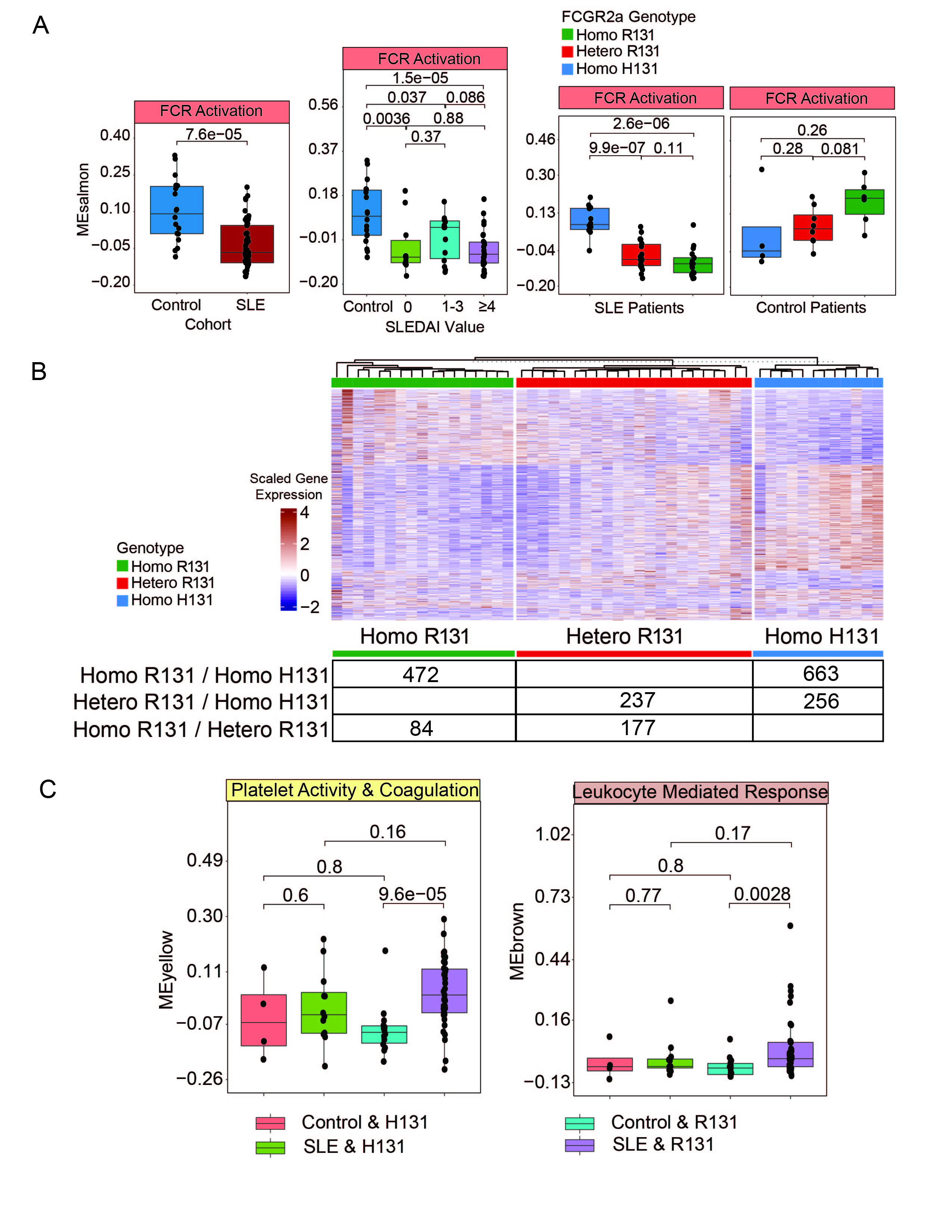
Results: Our cross-sectional SLE cohort (N=51, age = 41.1±12.3, 100% female, 45% Hispanic, 24% black, 22% Asian, 51% white, SLEDAI = 4.4±4.2) was comprised of patients consecutively enrolled excluding those on aspirin or anticoagulants. Compared to the 18 controls, there were 2290 (p.adj < 0.05) differentially expressed genes. (Figure 1 A,B) GSEA revealed positive enrichment for pathways related to interferon response, TNFa signaling, and coagulation in SLE. (Figure 1C) WGCNA was used to create a modular transcriptomic framework. (Figure 2A) Modules enriched for platelet activity, immune response, and WNT signaling were significantly increased in SLE versus controls. Moreover, modules enriched for interferon response and WNT signaling paralleled increases in disease activity. (Figure 2B) When analyzing patients with proteinuria, modules associated with oxidative phosphorylation and platelet activity were unexpectedly decreased. (Figure 2C) Analyzing the ratio of fold changes between SLE/Control vs SLE Proteinuria/SLE No Proteinuria, genes increased in SLE and those with proteinuria were enriched for immune effector processes, while genes increased in SLE but decreased in proteinuria were enriched for coagulation and cell adhesion. (Figure 2D) The module enriched for FCR activation was decreased in SLE and was affected by the FcγRIIa genotype. (Figure 3A) FcγRIIa R131 and H131 patients showed significantly different platelet transcriptomes. (Figure 3B) The combination of SLE with an FcγRIIa R131 variant leads to a significant increase in the platelet activity module not seen in controls. (Figure 3C)
Conclusion: These analyses reveal that SLE patients have a significantly different platelet transcriptome from controls, different phenotypic presentations of SLE patients associate with distinct platelet transcriptomic signatures, and FCGR2a variants may differentially influence the role of platelets in the contribution to SLE disease activity.
To cite this abstract in AMA style:
Cornwell M, EL Bannoudi H, Luttrell-Williams E, Myndzar K, Engel A, Izmirly P, Belmont H, Clancy R, Berger J, Ruggles K, Buyon J. Modeling of Clinical Phenotypes in SLE Based on Platelet Transcriptomic Analysis and FCGR2a Biallelic Variants [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/modeling-of-clinical-phenotypes-in-sle-based-on-platelet-transcriptomic-analysis-and-fcgr2a-biallelic-variants/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/modeling-of-clinical-phenotypes-in-sle-based-on-platelet-transcriptomic-analysis-and-fcgr2a-biallelic-variants/