Session Information
Session Type: Abstract Session
Session Time: 4:00PM-5:30PM
Background/Purpose: Juvenile Dermatomyositis (JDM) is a rare multisystem autoimmune condition that involves complex immune responses in both innate and adaptive compartments. Currently, we have a limited systems-level understanding of how cellular immunophenotypes functionally cooperate and are dysregulated in JDM. Increasing knowledge of the immune regulatory networks associated with JDM could lead to identification of precision treatment strategies and biomarkers.
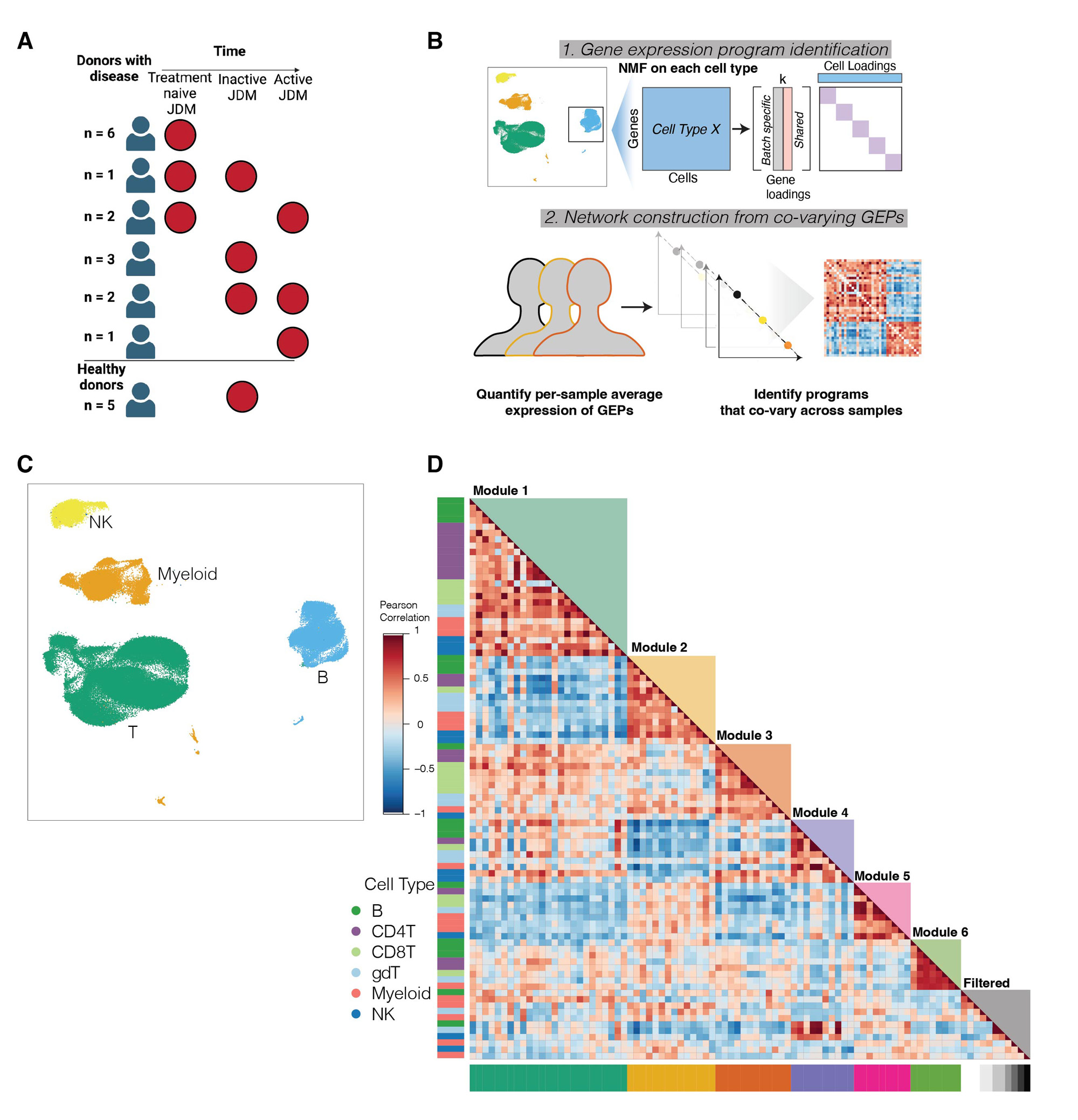
Methods: We used multiplexed single cell RNA sequencing to profile 27 peripheral blood samples from JDM patients (n=15, samples=22) and healthy pediatric controls (n=5, HCs). We performed standard processing, dimensional reduction, and clustering steps to identify 6 major cell types (B, CD4T, CD8T, gdT, NK & Myeloid). We applied an unsupervised network inference method, DECIPHERseq, which uses non-negative matrix factorization to identify coordinated regulatory networks of functional gene expression programs (GEPs) across cell types and community detection to identify hubs or ‘modules’ of GEPs. The network was annotated using 1) gene set enrichment analysis to identify pathways overrepresented in each program and 2) the DECIPHERseq enrichment method, which relies on resampling to identify gene sets enriched within modules compared to the rest of the network. To determine JDM disease activity-associated changes in network structure, we performed ANOVA on mean patient expression of GEPs between treatment-naive (TN), active, and inactive JDM, and HC samples.
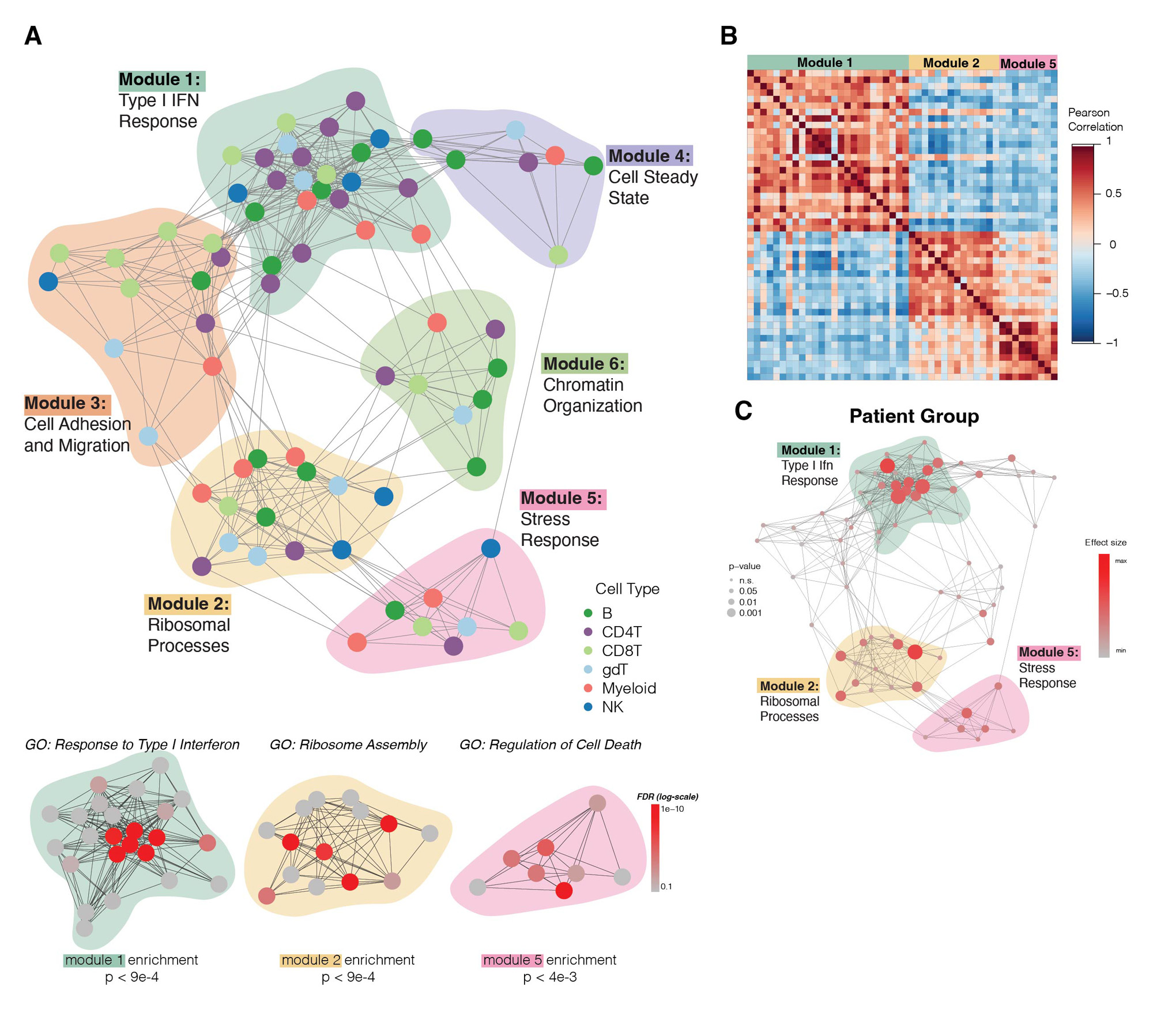
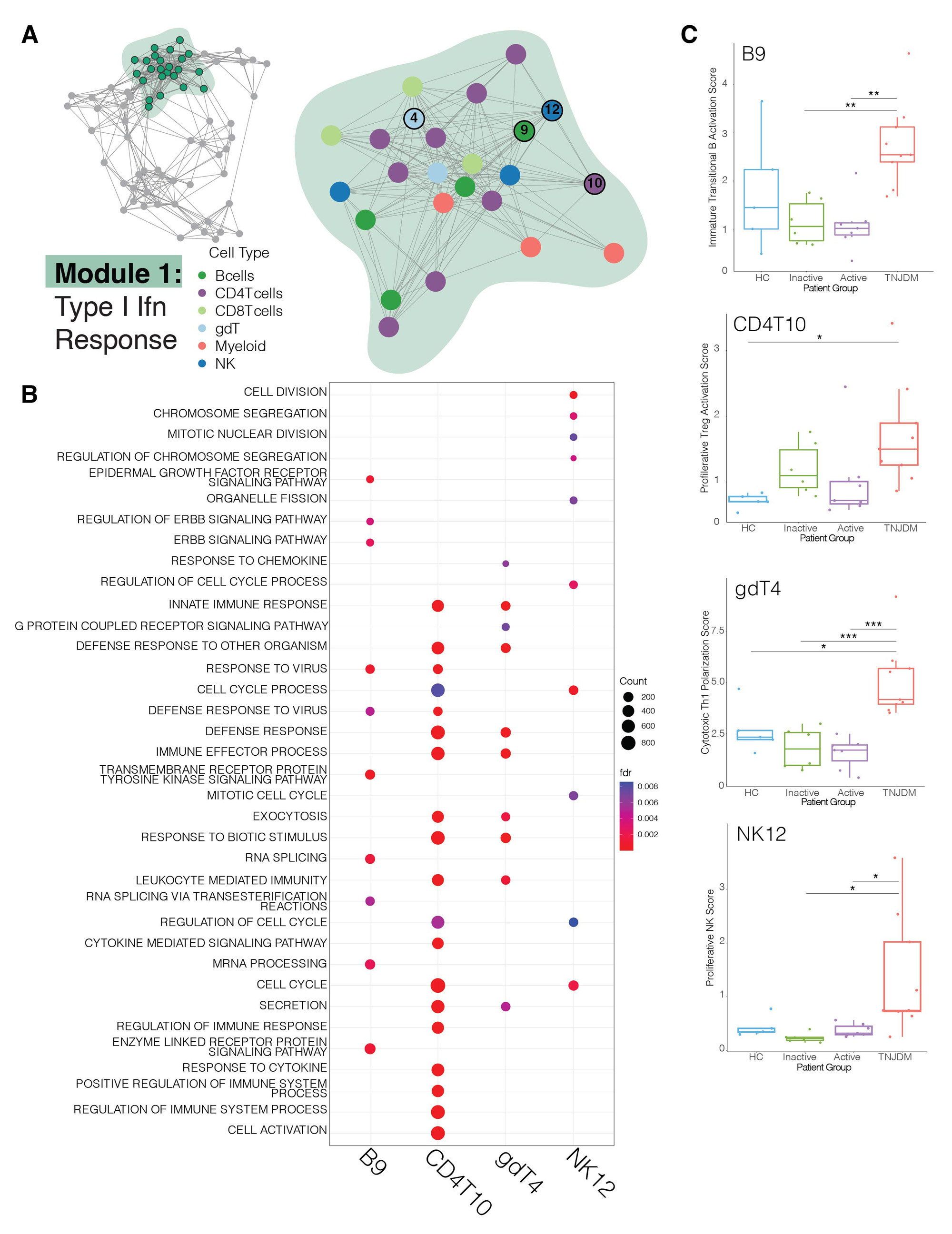
Results: We analyzed ~110,000 cells. DECIPHERseq inferred a network of 76 activity programs that formed 6 modules. All modules contained multiple cell types, highlighting that biological processes are coordinated across many cell types in JDM. Module enrichment analysis revealed consensus biological themes for each GEP hub. Module 1 was enriched in type I IFN responses and many programs in this module were increased in TN-JDM, as expected. Several disease-associated programs were highly correlated to this central IFN hub suggesting these cell-specific responses may be either up or downstream of IFN signaling. These included NK12, a proliferative NK program (MKI67, HIST1H1B); gdT4, a cytotoxic Th1 polarized gdT program (GZMB, CX3CR1, TBX21); CD4T10, an activated and proliferative Treg program (FOXP3, IL2RA, PRDM1, MKI67); and B9, an immature naïve B cell program (CD9, TCL1A, CD24), all of which were expressed higher in TN-JDM. Module 2, enriched in ribosomal processes, and Module 5, enriched in stress responses and regulation of cell death, were negatively correlated with Module 1.Several programs within Modules 2 and 5 were expressed significantly lower in TN-JDM suggesting dysfunction of these cellular processes in new-onset disease.
Conclusion: By employing unsupervised network analyses, we identified a central IFN hub highly correlated with novel cell-specific GEPs as well as dysfunction of ribosome and cell stress responses across many cell types in TNJDM. This systems-level perspective highlights coordinated peripheral immune responses that are both over- and underactive in JDM providing a foundation for future work to identify therapies to reprogram this dysregulation.
To cite this abstract in AMA style:
Rabadam G, Wibrand C, Flynn E, Hartoularos G, Sun Y, Kim S, Ye C, Gartner Z, Sirota M, Neely J. Mapping the Network of Coordinated Immune Dysregulation in Juvenile Dermatomyositis at Single-cell Resolution [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/mapping-the-network-of-coordinated-immune-dysregulation-in-juvenile-dermatomyositis-at-single-cell-resolution/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/mapping-the-network-of-coordinated-immune-dysregulation-in-juvenile-dermatomyositis-at-single-cell-resolution/