Session Information
Session Type: ACR Plenary Session
Session Time: 11:00AM-12:30PM
Background/Purpose: SLE is a systemic autoimmune disease that predominantly affects women in reproductive years. SLE pregnancies result in higher rates of adverse outcomes compared with healthy pregnancies.
Methods: To understand the molecular mechanisms underlying SLE pregnancy, we characterized the blood transcriptome of 135 pregnant (92 SLE and 43 healthy) subjects from the PROMISSE Study and 54 non-pregnant (NP) (20 SLE and 34 healthy controls) women by microarray. Blood was drawn at 4 time points during pregnancy (P1: <15 weeks gestation; P2: 16-23 wks; P3: 24-31 wks; P4: 32-40 wks) and 8-20 wks postpartum (PP). Poor pregnancy outcomes were classified as preeclampsia (SLE-PE) or other complications (SLE-OC) that included fetal or neonatal death, preterm delivery (<36 wks because of IUGR or placental insufficiency), and growth restriction (<5th %ile). SLE pregnancies included 24 SLE-PE, 22 SLE-OC, and 46 non-complicated (SLE-NC). Data were analyzed using a linear mixed model accounting for disease, complication groups and visit time point.
Results:
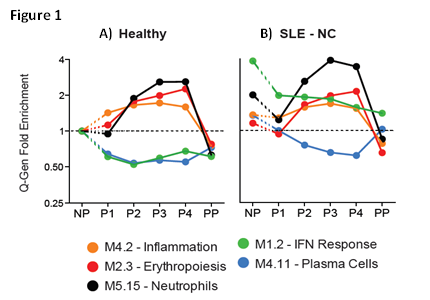
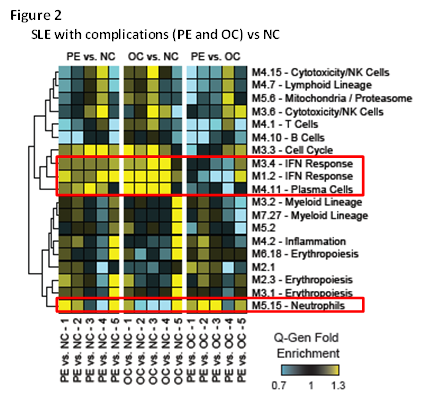
We first identified transcriptional signatures associated with healthy pregnancy. Using healthy NP controls as baseline, 9,687 transcripts were differentially expressed in healthy pregnancy; these included upregulation of neutrophil, myeloid inflammation and erythropoiesis signatures and downregulation of immune pathways linked to lupus pathogenesis, such as IFN and plasma cells (Fig. 1A). We then assessed how the signature was affected in SLE-NC compared to healthy NP women. SLE-NC pregnancies displayed the dynamic features similar to healthy pregnancies (Fig. 1B). However, while the plasma cell signature was decreased to levels lower than those of SLE-NP and healthy NP controls, the IFN signature remained patent through the course of pregnancy compared to healthy NP controls (green line). Finally, we identified complication-related transcriptional signatures by comparing SLE-PE and SLE-OC to SLE-NC (Fig. 2). Both SLE groups with complications failed to downregulate IFN and plasma cell signatures to SLE-NC levels throughout the pregnancy. In addition, SLE-PE displayed early upregulation of neutrophil signatures. Network analysis at P1 identified neutrophil- (AZU1, CTSG, ELANE) related signaling pathways as early biomarkers of preeclampsia.
Conclusion: Blood transcriptomics reveals systemic changes during healthy and SLE pregnancies and highlights the neutrophil signature as a potential early biomarker of preeclampsia.
To cite this abstract in AMA style:
Hong S, Banchereau R, Guerra MM, Salmon JE, Pascual V. Longitudinal Blood Transcriptomics Uncovers Immune Networks Associated with Complications in Lupus Pregnancy [abstract]. Arthritis Rheumatol. 2016; 68 (suppl 10). https://acrabstracts.org/abstract/longitudinal-blood-transcriptomics-uncovers-immune-networks-associated-with-complications-in-lupus-pregnancy/. Accessed .« Back to 2016 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/longitudinal-blood-transcriptomics-uncovers-immune-networks-associated-with-complications-in-lupus-pregnancy/