Session Information
Session Type: Poster Session D
Session Time: 8:30AM-10:30AM
Background/Purpose: LN is a severe consequence of SLE and there is a huge unmet need for discovery of urine protein biomarkers that provide non-invasive surrogates of disease activity and response to therapy. The purpose of this study was to measure protein biomarkers in urine samples from a diverse cohort of LN patients and to assess their correlations with patient demographics and clinical characteristics including the renal measures of estimated glomerular filtration rate (eGFR), SLEDAI-renal (SLEDAI-R) and National Institutes of Health Activity (NIH-AI) and chronicity (NIH-CI) indices
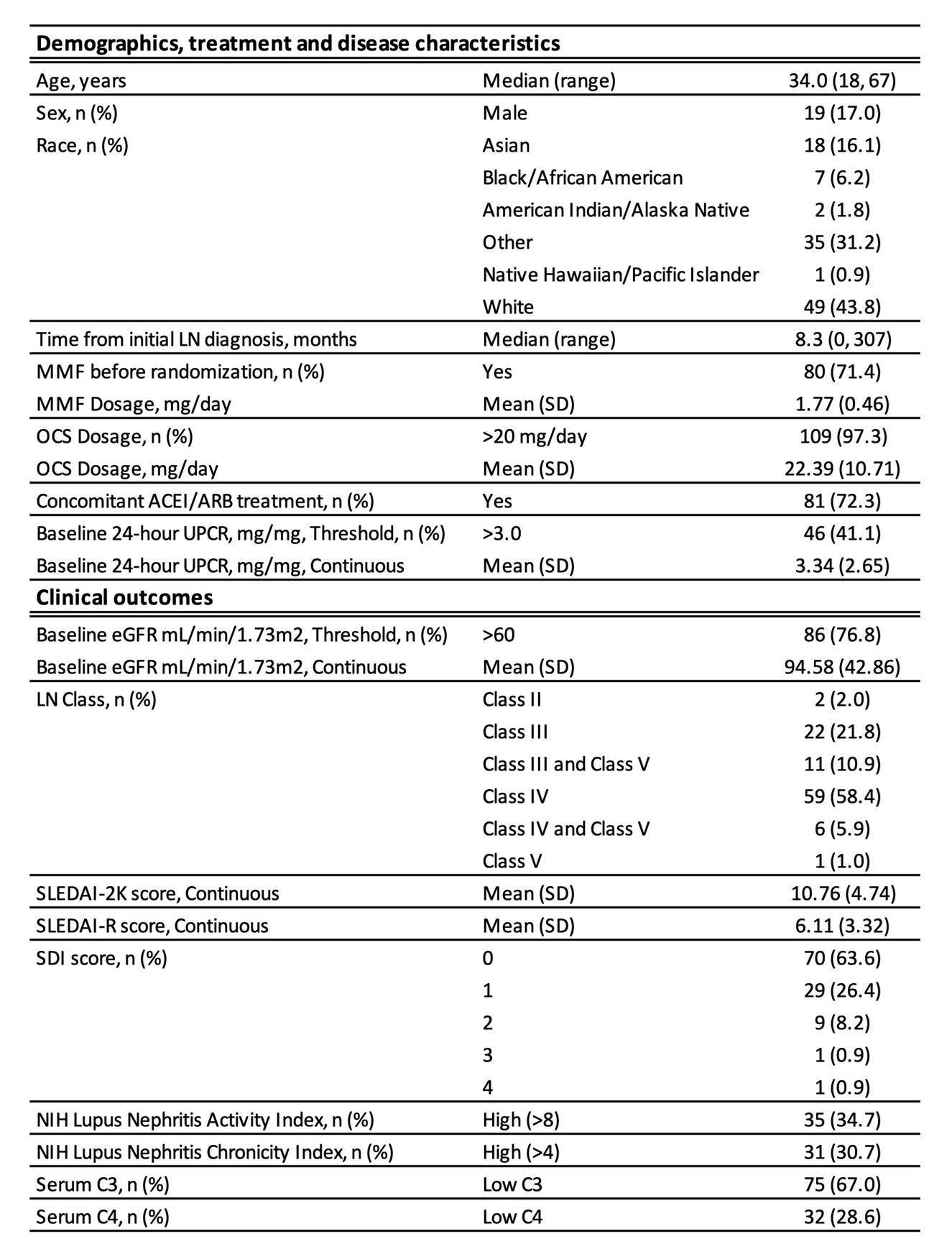
Methods: The demographics and characteristics of the cohort of 112 patients is described in Table 1. All patients fulfilled the 1997 ACR criteria for SLE and all had biopsy-proven LN. Urine samples from patients and 16 healthy donors (HD) were analyzed for 192 proteins by Luminex and 5 by SimoaTM. Protein concentrations were normalized to urinary creatinine levels. Log-concentrations of each protein was assessed for correlation with each clinical feature using linear regression adjusted for age, gender, ethnicity, disease duration, and treatment. eGFR was assessed using the cutoff of >60. The Benjamini-Hochberg procedure was used to calculate false discovery rates, with a cut-off of 0.1 used to determine significance. Statistical significance of intersections of protein lists was assessed via permutation. Pathway assessments were conducted using Ingenuity core analysis.
Results: Pre-filtering proteins for LN normalized mean concentrations to be greater than the HD mean + 1.5 SD yielded 97 distinct upregulated proteins. The largest numbers of protein-outcome associations were confounded by age (but not disease duration), gender, and MMF dose. After removing the influence of all confounders, numerous proteins showed statistically significant differential expression with respect to eGFR, SLEDAI-2K, SLEDAI-R, serum C3 and C4, NIH-AI, and NIH-CI. Conversely, no proteins were significantly correlated with LN class, race, or SDI (SLICC/ACR disease Index). The highest numbers of significant proteins were found for eGFR (55 proteins) followed by SLEDAI-R (36) and NIH-AI (20). There was a significant overlap of 11 proteins (Table 2) across these three lists (intersection p=4.1 × 10-5), indicating that these clinical renal measures share common biological processes. Pathway analysis of the 11 common proteins indicated enrichment for functions known to be associated with fatty acid/lipid metabolism, cardiovascular disease, cellular trafficking and immune cell infiltration
Conclusion: By analyzing LN urinary proteomics, we revealed that 3 clinical renal measures: eGFR, SLEDAI-R and NIH-AI are commonly associated with various proteins and pathways, most of which support the emerging importance of renal vascular pathology in LN.
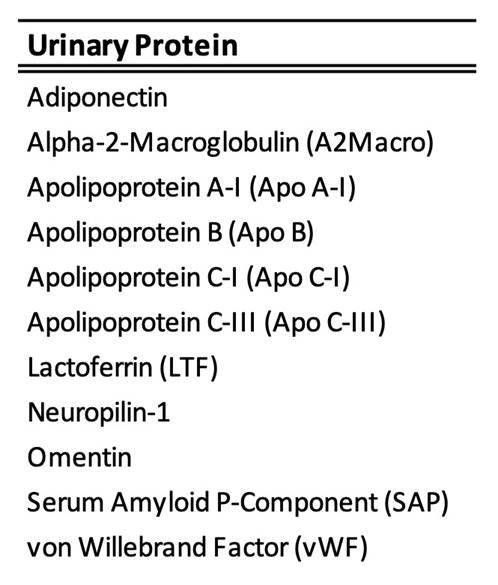 Table 2: 11 urinary proteins which reached statistical significance (false discovery rate < 0.1) for association with eGFR, SLEDAI-R and NIH-AI
Table 2: 11 urinary proteins which reached statistical significance (false discovery rate < 0.1) for association with eGFR, SLEDAI-R and NIH-AI
To cite this abstract in AMA style:
Newcombe P, Ramaswamy M, Sinibaldi D, Lindholm C, Jones F, Akhgar A, Brohawn P, Tummala R, White W. LN Urinary Proteomics Reveals Common Biological Pathways Identified by Distinct Disease Measures [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/ln-urinary-proteomics-reveals-common-biological-pathways-identified-by-distinct-disease-measures/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/ln-urinary-proteomics-reveals-common-biological-pathways-identified-by-distinct-disease-measures/