Session Information
Session Type: ACR Concurrent Abstract Session
Session Time: 4:30PM-6:00PM
Background/Purpose:
Recent studies suggest that epigenetic marks distinguish FLS isolated from different joints in RA. Hip and knee joint-derived FLS, in particular, have distinctive DNA methylation and transcriptome patterns that implicate the “IL-6 signaling pathway” and the “JAK-STAT pathway”. In this study, we determined how hip and knee FLS in RA differ in terms of the response to IL-6 to define the functional sequelae of joint-specific epigenetic imprinting.
Methods:
RA FLS lines from hip and knee arthroplasties were used from passage 5-7 (5 each). FLS were serum starved and then stimulated with IL-6 or medium for 2h and RNA-seq was performed. Differential gene expression was determined using edgeR. Gene set enrichment analysis was performed using MSigDB curated gene sets. Differentially modified epigenetic regions (DMERs) between hip and knee FLS were analyzed from published ATAC-seq and histone ChIP-seq using DiffBind (FDR<0.05). Published whole genome bisulfite sequencing (WGBS) datasets were analyzed using DSS. Western blot analysis and/or solid phase immunoassay were performed using antibodies to P-STAT3, STAT3, JAK1 and GAPDH.
Results:
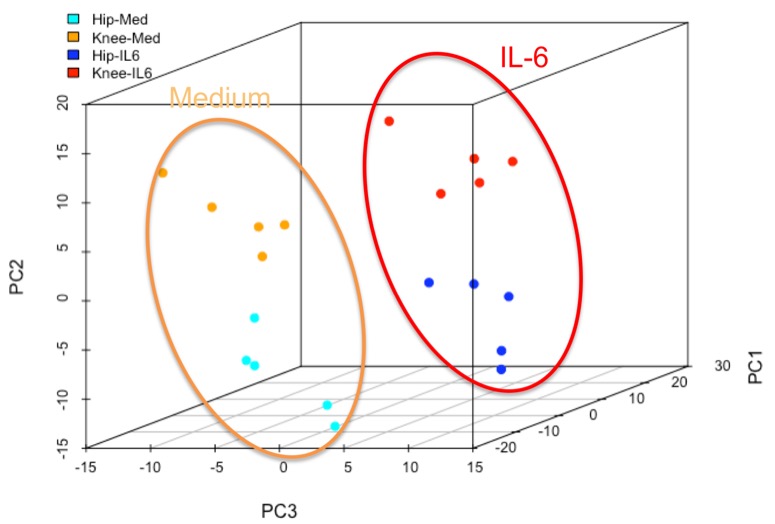
RNA-seq data were normalized and principal component analysis was performed (Fig 1). Unstimulated hip and knee FLS transcriptomes segregate, consistent with previous studies. The hip and knee FLS also segregated after IL6 treatment, indicating that joint-specific differences are maintained after IL6 treatment and do not converge. Differentially expressed genes (DEGs) were identified and we focused on hip vs knee differences present in both medium and IL6-treated FLS. Gene Set Enrichment Analysis indicated that the knee was enriched in genes in “Cell adhesion molecules” and “Integrin pathway”, while the hip was enriched in “p38_alpha beta downstream pathway”. Next, ATAC-seq, histone ChIP-seq and WGBS datasets from RA FLS were explored to identify additional epigenomic differences between the hip and knee in IL6-signaling. In unstimulated FLS, WGBS data showed 4 differentially methylated sites associated with JAK1 and 5 sites associated with gp130. ATAC-seq showed differences in chromatin accessibility in the IL6 receptor and JAK1. The histone H3K27ac was differentially marked for the STAT3 gene. To understand the mechanism of differential IL6 responses, we evaluated hip and knee RA FLS after IL6 stimulation. P-STAT3 levels peaked after 30min and returned to baseline at 4h. Knee FLS had significantly higher P-STAT3 than hip FLS at 30 minutes (1.2±0.4 vs 0.5±0.2, p<0.05), indicating that enhanced STAT activation accounts for some transcriptome differences between RA hip and knee FLS.
Conclusion:
RA hip and knee FLS have distinct transcriptomes, epigenetic marks, and STAT3 activation patterns in the IL6 pathway. These differences could contribute to joint-location responses to targeted therapies such as JAK inhibitors.
To cite this abstract in AMA style:
Hammaker D, Nygaard G, Kuhs A, Ai R, Boyle DL, Wang W, Firestein GS. Joint Location-Specific IL6 and JAK-STAT Signaling in Rheumatoid Arthritis (RA) Fibroblast-like Synoviocytes (FLS) [abstract]. Arthritis Rheumatol. 2018; 70 (suppl 9). https://acrabstracts.org/abstract/joint-location-specific-il6-and-jak-stat-signaling-in-rheumatoid-arthritis-ra-fibroblast-like-synoviocytes-fls/. Accessed .« Back to 2018 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/joint-location-specific-il6-and-jak-stat-signaling-in-rheumatoid-arthritis-ra-fibroblast-like-synoviocytes-fls/