Session Information
Date: Sunday, November 8, 2020
Title: T Cell Biology & Targets in Autoimmune & Inflammatory Disease Poster
Session Type: Poster Session C
Session Time: 9:00AM-11:00AM
Background/Purpose: Dermatomyositis (DM) is a systemic autoimmune disease affecting the skin and muscles, among other organs. The inflammatory infiltrate in skin has not been fully characterized, but the type I interferon (IFN) system is thought to be a major contributor to disease and has the capacity to interact with multiple cell types. Previous studies have used single or double staining to characterize the complex inflammatory infiltrate, but we implemented image mass cytometry (IMC) and took an unbiased approach to identify possible cellular sources of the type I IFNs and other cytokines.
Methods: We evaluated skin biopsies from 10 patients with moderate-severe DM and 5 healthy controls (HC). IMC was implemented to identify the infiltrate at a single cell level using a diverse set of 37 metal conjugated antibodies. Biopsies were stained with a cocktail of the 37 markers and regions of 1mm x 2mm ablated using the Hyperion Imaging System. Cell were segmented using a nuclear app-based algorithm in Visiopharm. Per cell mean pixel intensity (MPI) analysis was performed using histoCAT. The Phenograph algorithm was used to cluster cells based on expression of cell type markers. Further gating for rare populations was performed using FlowJo and image based sliding scale gates. Statistical analysis included the Mann-Whitney and Spearman rank correlation tests.
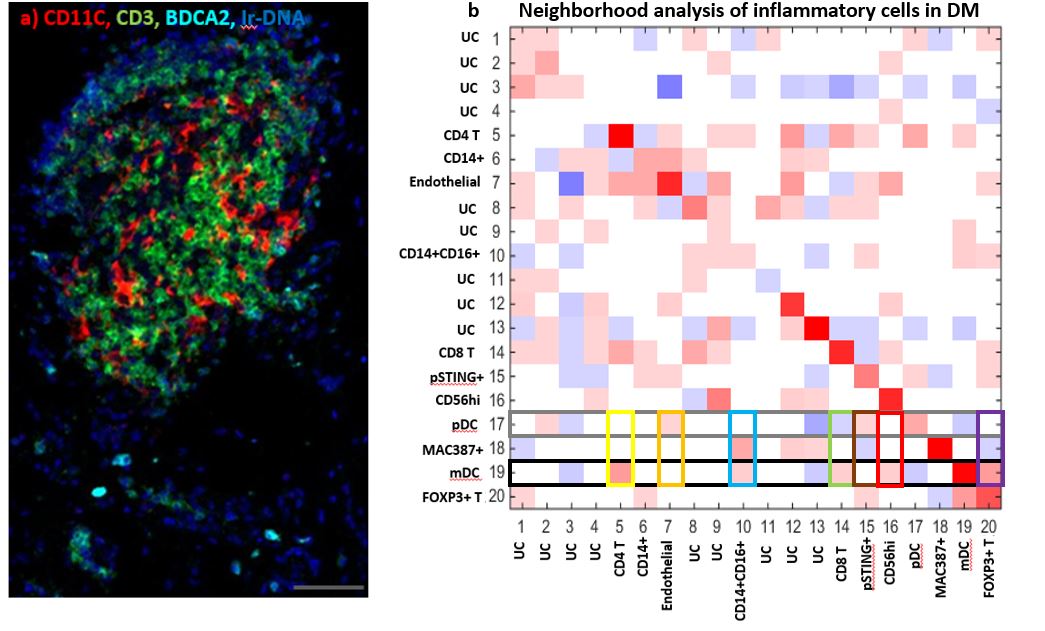
Results: We identified CD14+, CD14+CD16+, MAC387+, Endothelial, CD4 T, FOXP3+ T, Mast cells, CD56+, myeloid dendritic (mDC), plasmacytoid dendritic (pDC), CD8+ T, pSTING+, and CD20+ B cells (Fig 1). Monocyte-macrophage diversity was observed with increased numbers of CD14+, CD14+CD16+, MAC387+, and pSTING+ macrophages in lesional DM skin (p< 0.05), with the CD14+ population correlating positively with the cutaneous dermatomyositis disease area and severity index (CDASI) (r=0.697, p=0.031). The T cell compartment revealed CD4, CD8, and FOXP3+ T cells with activated CD69+ circulating memory T cells correlating with CDASI scores (r=0.754, p=0.0151). Inflammatory pathways pPPARγ, pSTING, IL31, IFNβ, pIRF3, pTBK1, IFNγ, IL4, and IL17 were all upregulated compared to HC (p< 0.05, Fig 2); pERK did not differ. IFNβ protein is upregulated in T cell, macrophage, dendritic, and endothelial cell populations of DM skin (p< 0.05) with mDC IFNβ trending highest. IL4+ and IL31+ mDCs were found to correlate with a Skindex itch score (IL4+: r=0.6547, p=0.0448; IL31+: r=0.7310, p=0.0194). pDCs colocalize with IFNγ in addition to the known IFNβ. The endothelium, mDCs, and B cells in DM skin express pPPARγ. Neighborhood analysis reveals significant interactions between mDCs and CD4, CD8, FOXP3+ T cells while pDCs interact with pSTING+ macrophages (p< 0.05, Fig 3).
Conclusion: The immune infiltrate in DM is complex and consists of a dominant monocyte-macrophage-myeloid cell axis that may contribute to IFNβ related pathogenesis and IL4, IL31 related pruritus. Findings of interest are CD14+ monocytes, pSTING+ macrophages, IFNγ+ pDCs, and pPPARγ+ endothelial cells and active mDCs, which requires further investigation. The use of IMC allows for single cell characterization of the immune cell population seen in DM skin and possible novel targeted therapeutics.
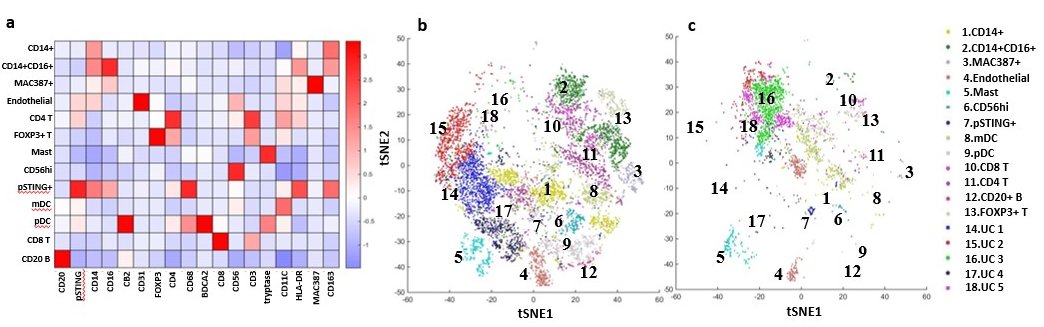 Immune Cells in Dermatomyositis Skin Lesions a) Heatmap of Different Cell Type Markers expressed the 13 cell populations identified: Monocytes (CD14+CD16-), mo-MAC (CD14+CD16+), Macrophages (MAC387+), Endothelial Cells (CD31+), CD4 T Cells (CD4+CD3+), Tregs (FOXP3+CD4+CD3+), Mast Cells (tryptase++), CD56+ Cells, pSTING+ mo-MAC (pSTING++,CD14+CD16+), mDC (CD11C+HLADR+), pDC (BDCA2+), CD8 T (CD8+CD3+), and CD20 B (CD20+). b) tSNE dimensional reduction plot of the different cell clusters identified in lesional DM skin c) tSNE dimensional reduction plot of the corresponding cell clusters in HC skin. Clusters 14-18 represent unidentified clusters (UC)
Immune Cells in Dermatomyositis Skin Lesions a) Heatmap of Different Cell Type Markers expressed the 13 cell populations identified: Monocytes (CD14+CD16-), mo-MAC (CD14+CD16+), Macrophages (MAC387+), Endothelial Cells (CD31+), CD4 T Cells (CD4+CD3+), Tregs (FOXP3+CD4+CD3+), Mast Cells (tryptase++), CD56+ Cells, pSTING+ mo-MAC (pSTING++,CD14+CD16+), mDC (CD11C+HLADR+), pDC (BDCA2+), CD8 T (CD8+CD3+), and CD20 B (CD20+). b) tSNE dimensional reduction plot of the different cell clusters identified in lesional DM skin c) tSNE dimensional reduction plot of the corresponding cell clusters in HC skin. Clusters 14-18 represent unidentified clusters (UC)
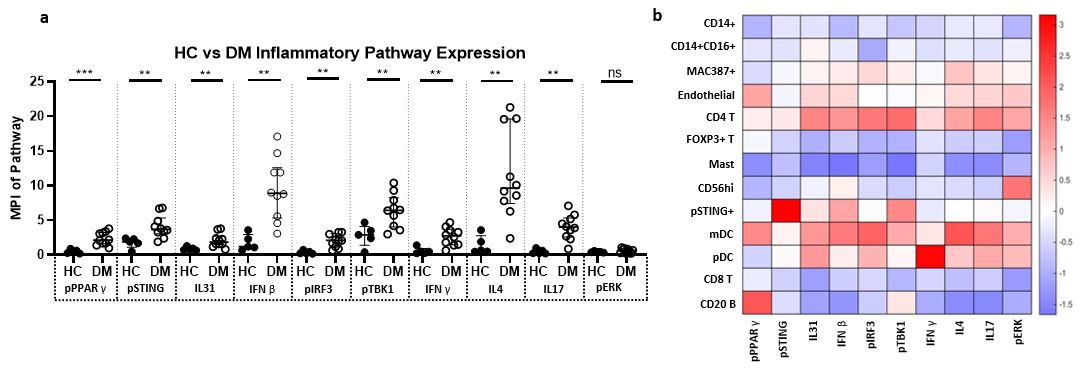 Inflammatory Pathway expression in Dermatomyositis Skin Lesions a) Expression of pPPARγ, pSTING, IL31, IFNβ, pIRF3, pTBK1, IFNγ, IL4, IL17 is increased in DM skin compared to HC skin. There is no significant difference between DM and HC skin pERK expression. b) Heatmap displaying the relative mean expression of each inflammatory pathway in each cell cluster * p < 0.05, ** p < 0.01, *** p < 0.001
Inflammatory Pathway expression in Dermatomyositis Skin Lesions a) Expression of pPPARγ, pSTING, IL31, IFNβ, pIRF3, pTBK1, IFNγ, IL4, IL17 is increased in DM skin compared to HC skin. There is no significant difference between DM and HC skin pERK expression. b) Heatmap displaying the relative mean expression of each inflammatory pathway in each cell cluster * p < 0.05, ** p < 0.01, *** p < 0.001
To cite this abstract in AMA style:
Patel J, Maddukuri S, Li Y, Bax C, Werth V. Investigating the Dermatomyositis Skin Inflammatory Infiltrate Using Image Mass Cytometry [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/investigating-the-dermatomyositis-skin-inflammatory-infiltrate-using-image-mass-cytometry/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/investigating-the-dermatomyositis-skin-inflammatory-infiltrate-using-image-mass-cytometry/