Session Information
Date: Tuesday, October 28, 2025
Title: (1830–1854) Systemic Lupus Erythematosus – Etiology and Pathogenesis Poster
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Preeclampsia (PE) is a serious complication of pregnancy associated with significant maternal and fetal outcomes. Systemic lupus erythematosus (SLE) significantly increases the risk of PE, which is not explained by traditional risk factors. A dysregulated immune response, with altered gene expression and aberrant immune cell activation play a critical role in the development of PE in patients with SLE. We conducted an integrative bioinformatics analysis to identify candidate genes and pathways implicated in PE among patients with SLE. By leveraging publicly available transcriptomic datasets, we aimed to uncover potential early biomarkers and explore their association with immune cell populations in peripheral blood, providing insights into the immunopathogenesis of PE in SLE.
Methods: We selected the next datasets from the Gene Expression Omnibus database: GSE177029, GSE185047, and GSE48424 (Table 1). We filtered differentially expressed genes (DEGs) using the R-based tool GEO2R, with a false-discovery rate (FDR) value of < 0.05 and a log2foldchange >1 for upregulated genes. A Venn diagram was drawn to visualize shared DEGs across datasets. Enrichment analysis was performed to retrieve gene ontology (GO) biologic processes (BP) and KEGG pathways using ShinyGO. Protein-protein interactions (PPIs) were analyzed with the Cytoscape software, and key hub genes (HGs) were identified with the Cytohubba plugin using the maximal clique centrality algorithm. The Cibersort online tool was utilized to characterize the immune cell population from peripheral blood in dataset GSE48424. A two-way ANOVA test was done to compare immune cell proportions. The two-stage step up method was applied as a post-hoc test to control the FDR. Mean fluorescence intensity (MFIs) values were retrieved, and a Spearman correlation test was performed to assess the relationship between the HG expression and immune cell populations.
Results: 40 DEGs were shared in at least two different datasets (Figure 1A), and whose functional enrichment was notable for BPs related to regulation of viral genome replication and response to type I interferon (Figure 1C). Such terms were then supported by KEGG pathways related to viral infections such as pertussis, measles and influenza A (Figure 1D). PPI analysis successfully depicted 40 nodes and 172 edges. The MCC algorithm identified the top 10 HGs (Figure 1B) Regarding immune cell populations in peripheral blood, significant differences were observed in CD8+ T cells and neutrophils proportions. Finally, IFIT1, OAS3, IFI27, LY6E, OAS1, IFI44L, XAF1, and ISG15 were observed to have significant correlations with different immune cells (Figure 2).
Conclusion: This study aimed to identify candidate early biomarkers for preeclampsia in pregnant patients with SLE with novel approach. Ten candidate genes were identified and showed to be significantly correlated with immune cell populations in peripheral blood in patients with preeclampsia. These results provide a molecular framework for the study of preeclampsia in pregnant patients with SLE, which warrant further validation in a longitudinal cohort of patients.
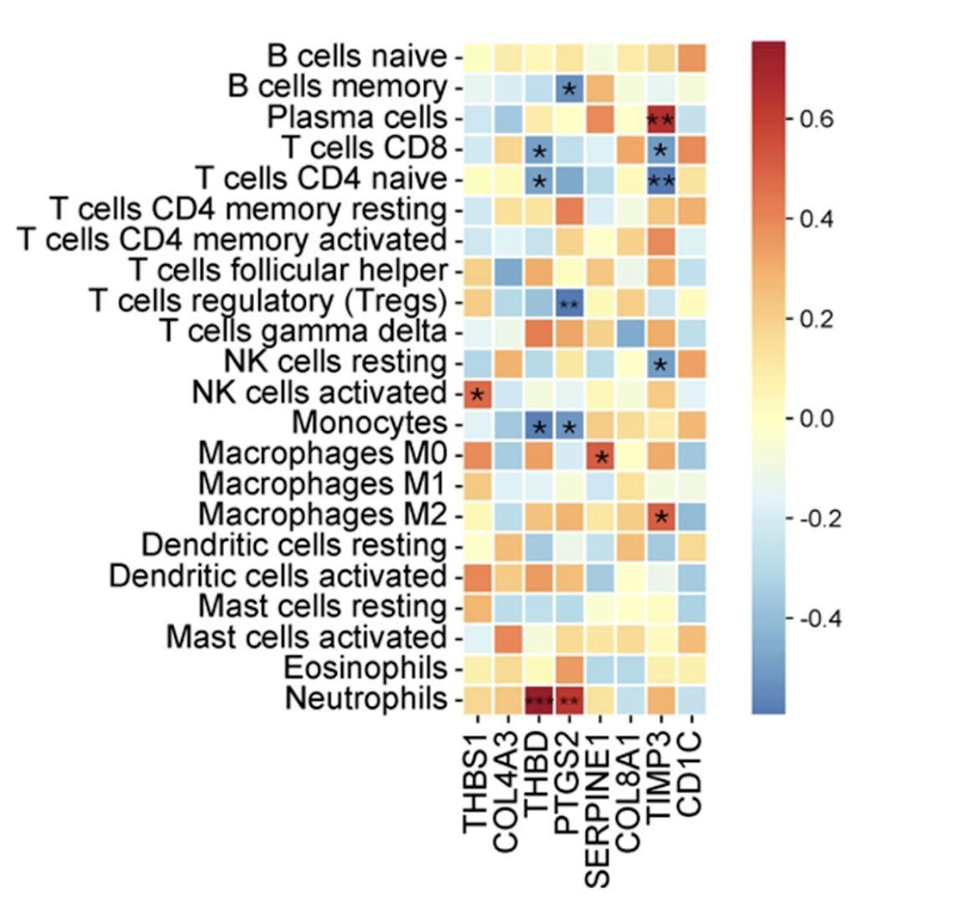 Figure 1. Correlation heatmap between candidate hub genes and immune cell populations in peripheral blood of pregnant women with systemic lupus erythematosus (SLE).
Figure 1. Correlation heatmap between candidate hub genes and immune cell populations in peripheral blood of pregnant women with systemic lupus erythematosus (SLE).
Spearman correlation coefficients are represented by color scale, with red indicating positive and blue indicating negative correlations. Significant correlations are marked with asterisks (*p < 0.05, **p < 0.01). Genes along the x-axis correspond to top hub genes identified through PPI analysis (e.g., THBS1, COL4A3, THBD), and immune cell subsets along the y-axis were estimated using CIBERSORT. Notable associations include negative correlations between THBD and neutrophils, and positive correlations between THBD and regulatory T cells (Tregs), suggesting immune regulatory involvement in preeclampsia pathogenesis in SLE.
.jpg) Figure2. Transcriptomic analysis of shared differentially expressed genes (DEGs) in preeclampsia among pregnant women with systemic lupus erythematosus (SLE). (A) Venn diagram showing overlapping DEGs among three GEO datasets: GSE177029, GSE185047, and GSE48424. A total of 40 genes were shared by at least two datasets. (B) Protein-protein interaction (PPI) network of the top hub genes identified via CytoHubba’s MCC algorithm in Cytoscape. Key interferon-stimulated genes such as IFIT1, OAS3, ISG15, and IFI44L were prominent. (C) Gene ontology (GO) enrichment analysis (biological processes) revealed significant overrepresentation of terms related to type I interferon signaling, viral genome replication, and immune response regulation. (D) KEGG pathway enrichment analysis identified key pathways involving infectious and inflammatory responses, including pertussis, measles, COVID-19, complement and coagulation cascades, and IL-17 signaling, highlighting immune activation as a central mechanism in SLE-associated preeclampsia.
Figure2. Transcriptomic analysis of shared differentially expressed genes (DEGs) in preeclampsia among pregnant women with systemic lupus erythematosus (SLE). (A) Venn diagram showing overlapping DEGs among three GEO datasets: GSE177029, GSE185047, and GSE48424. A total of 40 genes were shared by at least two datasets. (B) Protein-protein interaction (PPI) network of the top hub genes identified via CytoHubba’s MCC algorithm in Cytoscape. Key interferon-stimulated genes such as IFIT1, OAS3, ISG15, and IFI44L were prominent. (C) Gene ontology (GO) enrichment analysis (biological processes) revealed significant overrepresentation of terms related to type I interferon signaling, viral genome replication, and immune response regulation. (D) KEGG pathway enrichment analysis identified key pathways involving infectious and inflammatory responses, including pertussis, measles, COVID-19, complement and coagulation cascades, and IL-17 signaling, highlighting immune activation as a central mechanism in SLE-associated preeclampsia.
To cite this abstract in AMA style:
Martinez-Canales R, Galindo-Calvillo E, Ortiz-Rios A, Avalos-Garcia B, Perez-Barbosa L, Galarza-Delgado D, Skinner-Taylor C, Salinas-Carmona M, Macias-Segura N. Integrative Bioinformatics Analysis Reveals Key Genes and Immune Profiles Associated with Preeclampsia in Lupus Pregnancy [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/integrative-bioinformatics-analysis-reveals-key-genes-and-immune-profiles-associated-with-preeclampsia-in-lupus-pregnancy/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/integrative-bioinformatics-analysis-reveals-key-genes-and-immune-profiles-associated-with-preeclampsia-in-lupus-pregnancy/

.jpg)